Abstract.
Chronic kidney disease (CKD) is characterized by a progressive loss of renal function over time. Histopathological analysis of the condition of glomeruli and the proximal convolutional tubules over time can provide valuable insights into the progression of CKD. Optical coherence tomography (OCT) is a technology that can analyze the microscopic structures of a kidney in a nondestructive manner. Recently, we have shown that OCT can provide real-time imaging of kidney microstructures in vivo without administering exogenous contrast agents. A murine model of CKD induced by intravenous Adriamycin (ADR) injection is evaluated by OCT. OCT images of the rat kidneys have been captured every week up to eight weeks. Tubular diameter and hypertrophic tubule population of the kidneys at multiple time points after ADR injection have been evaluated through a fully automated computer-vision system. Results revealed that mean tubular diameter and hypertrophic tubule population increase with time in post-ADR injection period. The results suggest that OCT images of the kidney contain abundant information about kidney histopathology. Fully automated computer-aided diagnosis based on OCT has the potential for clinical evaluation of CKD conditions.
Keywords: optical coherence tomography, chronic kidney disease, proximal convolutional tubule, medical image processing
1. Introduction
Chronic kidney disease (CKD) is a medical condition characterized by the progressive loss of the kidney’s function.1 Nowadays, 26 million U.S. patients suffer from CKD condition while millions more are potentially at risk.2 In estimate, more than half a million U.S. residents have end-stage renal disease (ESRD), which is associated with high mortality rates (163.8 deaths/1000 patients/year) and huge economic burdens ().2 CKD is classified into five stages of increasing severity.3 ESRD is the last stage of CKD when dialysis or transplant is needed to stay alive. The stages of CKD or the progressive loss of kidney function is mainly based on measured or estimated glomerular filtration rate (eGFR).4 eGFR is obtained by blood test, including serum creatinine level, together with age, sex, and sometimes other information.4 Pathology can provide additional information about the microstructure of declined kidney function by viewing tubular atrophy, glomerulosclerosis, and interstitial fibrosis. However, there are significant artifacts associated with excision biopsies and immersion fixation procedures.
Optical coherence tomography (OCT)5–8 has the advantage of allowing the analysis of the microscopic structure of kidney in a nondestructive manner. OCT has been used to image thermal tissue damage to the rat kidney resulting from laser ablation.9 OCT’s capability to resolve renal corpuscles and uriniferous tubules was first demonstrated on rat kidney ex vivo using high-resolution time-domain OCT.8 With the advent of high-speed Fourier-domain OCT technology, three-dimensional (3-D) imaging of renal microanatomy in vivo was enabled.10 The kidney microstructures prior to, during, and following exposure to renal ischemia can be observed in real time. In a recent study, Andrews et al.11 utilized OCT to visualize the characteristic histopathologic changes on aging rat kidneys in vivo. With the onset of severe proteinuria at 10 to 12 months of age, OCT reveals tubular necrosis/atrophy, interstitial fibrosis, tubular dilation, and glomerulosclerosis.11 With a further deterioration in kidney function at 16 to 18 months of age as indicated by rising creatinine levels, OCT reveals more extensive interstitial fibrosis and tubular atrophy, increased tubular dilation with cyst formation and more sclerotic glomeruli.11
Wierwille et al.12 investigated the feasibility of Doppler OCT to image kidney microcirculation, specifically, glomerular blood flow. Normal blood flow as well as the effects of acute mannitol and angiotensin II infusion has been observed and quantified. Using ultrahigh-sensitive optical microangiography, Zhi et al.13 demonstrated highly sensitive imaging of renal microcirculation in vivo. Normal peritubular capillary microcirculation as well as the changes in response to renal ischemia and reperfusion can be monitored and quantified using this method.
Since OCT has deeper penetration depth than confocal microscopy, it is able to penetrate the renal capsule surrounding human kidneys thereby enabling the characterization of renal tubules, glomeruli, and cortical blood vessels in human kidneys.14–16 Using image-processing algorithms, key microstructural parameters, such as tubular lumen diameter, can be automatically quantified.16 In vivo imaging of human kidney has been reported during kidney transplantation procedures,17 and clinical studies have indicated that the openness of tubular lumens observed by OCT has a strong correlation with the posttransplant recovery of renal function.18 These results suggest that OCT may be a useful tool in intraoperative monitoring and evaluation of transplant kidneys for predicting postgraft function after ischemia-reperfusion injury. OCT has been applied to other clinical studies, including assessing the morphological features of the endothelial and vascular injury induced by catheter-based renal nerve ablation,19 and differentiating between normal renal parenchyma and renal cell carcinoma.20,21
Computer-aided diagnosis (CAD) systems can interpret medical images and provide an advisory diagnosis decision for medical doctors. Those systems aim to both decrease the diagnosis error rate and reduce decision making time.22 A great amount of research in CAD systems has been conducted for different types of imaging modalities and imaging targets. CAD computation pipelines often include region proposing, feature extraction, and classification. Liu et al.23 detected epidural masses on CT scans by region proposing with -mean clustering, extracting a collection of texture features, and trained a support vector machine (SVM) classifier based on the texture features. Farag et al.24 detected an entire pancreas organ location in CT scans by region proposing with simple linear iterative clustering (SLIC) algorithm, extracted dense scale-invariant feature transform (dSIFT) features, and trained a random forest classifier based on dSIFT features. Jerebko et al.25 detected colonic polyps in CT scans by region proposing with manual seed selection, extracted 12 hand-coded features, such as area, sphericity, surface pixel intensity mean, and trained a neural network classifier based on these 12 features. For OCT-related CAD system, Wan et al.26 detected and classified breast tissues in OCT scans using a sliding window technique for region proposing, extracted local binary pattern (LBP) texture, and trained a neural network classifier based on the LBP texture feature vector. Qi et al.27 detected and classified dysplasia in Barrett’s esophagus in endoscopic OCT scans using a semiautomatic segmentation process based on global threshold and extracted six texture features. They used principal component analysis (PCA) to obtain the top two significant feature spaces and found the classification threshold from the receiver operating characteristic curve generated from 100 annotated training images. Qi et al.28 detected colonic crypt morphology in en-face image from OCT 3-D scan using semiautomatic marker-based watershed segmentation, extracted six region features, such as area, density, and eccentricity, and used PCA to obtain the top two significant feature spaces.
In recent years, deep convolutional neural network (ConvNet) has been increasingly adopted by CAD applications. With proper ConvNet architecture, the original computation pipeline of CAD, which consists of region proposing, feature extraction, and classification, can be condensed into region proposing and ConvNet classification or even simply a ConvNet that is trained to calculate both the class and bounding box of the object. Roth et al.29 detected a lymph node from the CT scan using a preliminary software to generate 3-D lymph node candidate segments and a five-layer ConvNet to classify the 3-D segments based on two-dimensional (2-D) image generated from a random viewpoint of the 3-D segments. Roth et al.30 detected a pancreas in CT scans using SLIC algorithm for region proposing and deep ConvNet to classify the SLIC segmentation. Liu et al.31 detected colitis in CT scans using a “selective search” algorithm for region proposing, a pretrained Alexnet on PASCAL 2007 dataset (natural images instead of medical images) for feature extraction, and an SVM for classification. Esteva et al.32 classified 757 types of skin cancer captured on an RGB camera using Google Inception ConvNet architecture that was end-to-end trained on 1.4 million annotated skin-cancer images.
In this study, we used OCT to evaluate CKD in a murine model induced by intravenous Adriamycin (ADR) injection into Munich-Wistar rats.33 We present a CAD system based on ConvNet to automatically detect and quantify tubular diameter and hypertrophic tubule population from OCT images at several post-ADR induction time points.
2. Method
2.1. Animal Models and Experimental Protocols
The animal protocol has been approved by the committee on animal care and use at the University of Maryland, College Park. A murine model of CKD was induced by injection of ADR () into the tail vein of Munich-Wistar rats.33 Once a week during the entire study protocol of 8 weeks, rats were weighed. The 24-h urine volumes were collected in metabolic cages. Fresh urine samples were tested for albuminuria (Albustix), and blood samples taken from the tail vein were analyzed for serum creatinine and BUN values (Beckman Coulter Creatinine and BUN Analyzers). In each week for the 8-week period, two rats were anesthetized with (4% induction, 1.5% during operation, ). The abdominal cavity was opened through a midline incision, and the left kidney was exposed and imaged using OCT. Following in vivo OCT evaluation, the kidneys were fixed in situ by flushing with warm (i.e., 37°C) oxygenated saline, followed immediately by phosphate buffered 2% paraformaldehyde and 0.1% glutaraldehyde. The fixed kidneys were excised and the rat euthanized by intracardiac injection of pentobarbital sodium to induce cardiac arrest. Blocks of fixed kidneys were embedded in paraffin, sectioned, and stained with hematoxylin and eosin.
2.2. Optical Coherence Tomography
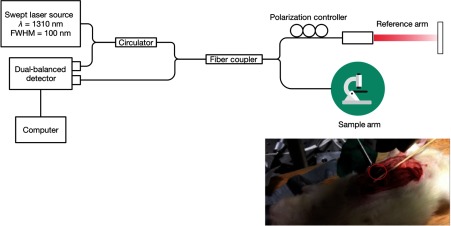
A custom-built Fourier-domain OCT system was used in this study (Fig. 1).34–37 This system uses a swept-source laser operating at 1310-nm center wavelength with 100-nm bandwidth. The axial and lateral resolutions of the system are 12 and , respectively. OCT image dimensions are 1024 pixels () in lateral direction and 512 pixels () in axial direction. The sensitivity of the system is 90 dB. The A-scan acquisition speed is 16 kHz. The 2-D image acquisition speed is 16 frames per second. For each experiment, 3-D OCT volumes were acquired from 5 to 10 kidney locations. Each volume consists of 475 consecutive 2-D images.
Fig. 1.
The schematic diagram of the custom-built OCT system used in this study. Inset image shows the abdominal cavity of a Munich-Wistar rat opened through a midline incision and the exposed left kidney for OCT imaging.
2.3. Computer-Aided Diagnosis Software
About 3000 to 5000 OCT kidney images have been acquired each week for eight weeks. To determine CKD condition from the massive image dataset, an OCT image analysis software is developed to automatically detect and measure features that are related to CKD progression. In our application, possible features can be the proximal tubule morphology, such as tubular diameter and atrophy/hypertrophy.38 The image analysis software consisted of four sections: image preprocessing, feature region proposing, feature region classification, and feature region measurement.
2.4. Image Preprocessing
Gaussian blur is used to reduce the speckle noise of the OCT image. Gaussian blur is performed by convoluting the original OCT image with a 2-D Gaussian kernel. As a low-pass filter, Gaussian kernel removes high-frequency noise as well as useful high-frequency information, such as edges. A small Gaussian kernel () is used to limit the blur effect and retain small and fast-varying features as much as possible.
2.5. Feature Region Proposing
To recognize multiple objects in the image, regions of interest (ROIs) have to be proposed to an image classifier. One of the easiest ways to propose ROIs is the sliding window algorithm. Specifying the range of window size and the stride size, a window slides from the edge of the image and proposes each window patch as an ROI. This method has been used widely and successfully in applications such as face recognition.39 However, as a greedy search method, it proposes a large number of proposed regions and suffers from “curse of dimensionality.”40 Sliding window method has to be combined with a very fast image classifier for speed-critical applications.
Another region proposing method is segmentation. Segmentation divides the image into subimages based on low-level image properties, such as pixel intensity and texture. For OCT images, image intensity-based segmentation with a global threshold cannot recall tubule lumens with high accuracy due to the nonuniform image brightness. A way to overcome this issue is to remove the effect of illumination in preprocessing, such as by the Retinex theory.41 However, in practice, those enhancement algorithms are very slow. In addition, the illumination effect may not be removed perfectly. Another way is to use dynamic local intensity thresholding. “Bradley” adaptive thresholding technique uses a computationally efficient integral image algorithm to determine the local threshold value for each subimage window.42 In comparison to other dynamic thresholding algorithms, Bradley adaptive thresholding is robust and less computationally expensive.43 A representative result of an OCT image segmented by Bradley adaptive thresholding is shown in Fig. 2(a).
Fig. 2.
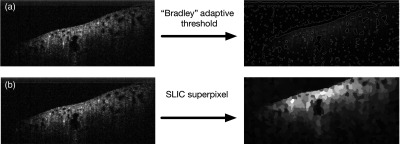
Same OCT image processed by (a) dynamic local intensity thresholding and (b) SLIC superpixel.
Superpixel is another region proposing method that groups similar pixels into a “superpixel.” Those superpixels can then be proposed as ROIs. The state-of-the-art superpixel methods include graph-based algorithm,44 gradient-ascent-based algorithm,45 and SLIC algorithm.46 A representative result of an OCT image processed by SLIC superpixel method is shown in Fig. 2(b). The principle of SLIC superpixel algorithm is the following: (1) first, equally spaced pixels are initialized as cluster centers, (2) each pixel then associates itself with the most similar neighboring cluster centers, (3) the cluster centers are updated to be the average pixel location of all its associated pixels, (4) repeat steps 1 to 3 until the new cluster center and the old cluster center converges. The number of cluster centers controls the granularity of superpixels or the total number of proposed regions.
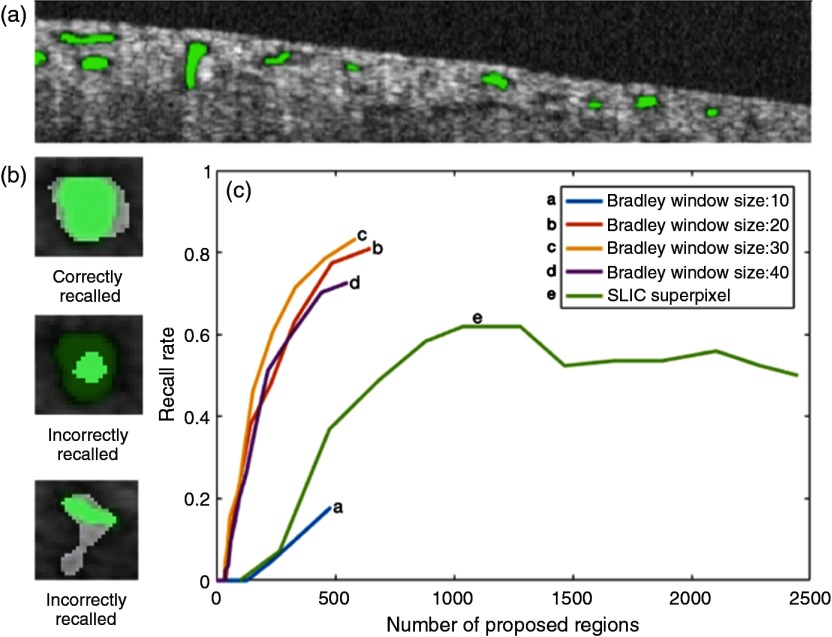
Ideally, the region proposing algorithm should have a 100% recall rate. Recall rate is defined as the percentage of total tubule lumens being proposed. Another key parameter is the total number of proposed regions. For example, sliding window algorithm with a small stride size and a large range of scale would have a near 100% recall rate, because it can theoretically capture all tubule lumens in different scales and at different locations. However, its total number of proposed regions is massive, which is impractical for many applications.
We compared the ROI recall rate and the total number of proposed regions between Bradley segmentation and SLIC superpixel. To determine the recall rate, tubule cross-sectional regions in test images are manually labeled [Fig. 3(a)]. A tubule region is correctly recalled if more than 60% of the recall region overlaps with the manually labeled region [Fig. 3(b)]. The number of proposed regions can be altered by changing the threshold level and window size for Bradley adaptive threshold algorithm or the initial number of cluster centers for SLIC superpixel algorithm. The recall rate versus the number of proposed regions for each algorithm was measured [Fig. 3(c)]. From the results, Bradley adaptive thresholding with a window size of 30 pixels is able to recall more than 80% of the tubule regions with under 500 proposed regions. SLIC superpixel algorithm underperforms Bradley adaptive thresholding for this application. Therefore, we choose Bradley adaptive thresholding with a window size of 30 pixels in this study.
Fig. 3.
(a) Manually labeled tubule lumens (green regions), (b) overlaps between proposed regions (white) and manually labeled true regions (green), and (c) recall rate versus number of proposed regions for different algorithms.
After candidate regions for proximal convolutional tubule lumens are chosen, rectangular image patches containing the candidate regions are cropped for further image classification. It is unavoidable that the set of proposed images would include many false positive regions, such as defects in the OCT image. From the perspective of adaptive threshold, they are similar to tubule cross sections, which have a relative darker center than the surrounding regions. Examples of the region-proposed images are shown in Fig. 4. To differentiate them, an image classifier is necessary to distinguish the true tubule cross sections [Fig. 4(a)] from the falsely proposed regions [Fig. 4(b)].
Fig. 4.
Examples of proposed region images: (a) tubule lumens and (b) OCT image artifacts and speckle noises. The brightness of some images has been increased for the ease of viewing. Sizes of the proposed region images range from 5 to 60 pixels in width.
2.6. Feature Region Classification: Convolutional Neural Network Image Classifier
There are many image classifier models. Some of the well-known classifiers include hard-coded classifier, eigen features classifier,47 and artificial neural network,48 etc. A classifier generated through learning tends to be more accurate due to the amount of prior information gained through the training process. In our study, OCT images contain large variations in tubule morphology, kidney condition, image quality, etc. ConvNet49 is eventually chosen as the image classifier due to its larger learning capacity.
ConvNet training requires thousands to millions of labeled images per detection class depending on the complexity of the network structure. In general, a large and complex network is capable of learning to classify more complicated visual features. However, it would also require a large annotated training database. Overfitting happens if a large network is trained with a small database, which leads to low-classification accuracy in testing. Therefore, there is a trade-off between learning capacity (network size) and training database size. In practice, despite the fact that sufficient image data are usually available, for a supervised learning model, such as ConvNet, it is usually the labor cost for annotating the thousands to millions of images that make creating a large training database a prohibitive task.
For our application, we built a training database with two classes: tubule class and nontubule class. To crop and collect training images, we first created a less-accurate image classifier based on hard-coded features. Bootstrapping from this classifier, we were able to collect sufficient images per class easily but with significant false classification rate. The incorrectly classified images in both classes were manually removed from the database by inspection. To keep the manual inspection time minimal, we deliberately collected only 2000 training images per each class. However, 2000 images per class are far from enough to train a network that has thousands of weights and biases without overfitting. To increase the database size, each of the original images was duplicated times. Then, the duplicated images went through random translation, rotation, scaling, and noise addition (Fig. 5). The database size was artificially increased by times. In our case, equals to 10 and the final database size is 20,000 images per class.
Fig. 5.
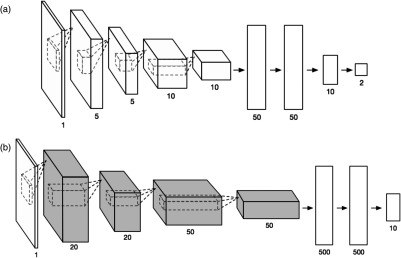
Multiple training images generated by manipulating the original tubule cross-section image (first on the left). All images are resized to for ConvNets training.
We attempted to train two different ConvNets with the database. The first network has a small structure size (less than the overfitting limit of the database) as shown in Fig. 6(a). The second network is the LeNet48 [Fig. 6(b)]. LeNet is chosen, because it is originally designed for recognizing single channel grayscale handwritten-digit images (MNIST dataset), which is in the similar format as OCT images. Many latest ConvNet architectures, such as AlexNet, are originally designed for RGB image classification; therefore, they are not included in this study. The smaller-sized network was trained end to end from the first to the last layer with each layer initialized randomly. LeNet was trained with transfer learning; instead of end-to-end training, it started from a pretrained LeNet for handwritten-digit recognition. Only the fully connected layers were fine-tuned using our training database. Transfer learning enables a large network to be trained on a smaller dataset without overfitting. The performance of the two networks was benchmarked with a separate testing database; the testing database contains OCT images randomly chosen from all CKD stages. Locations of the tubules in the testing database were manually labeled. The bench test result showed a better tubule recognition rate (90%) from LeNET with transfer learning. It is worth noting that whether transfer learning is a good option can be speculated based on the degree of visual differences between the original application of the pretrained network and the current application.50 The ConvNet training was performed using MatConvNet51 on MATLAB® using an Nvidia GTX660 GPU. Once the network was trained, the network weights were saved. Image classification based on the weights could be performed on machines without Nvidia GPU support. In our study, the image classification based on the trained ConvNet was programmed in C++, with multithreading parallelism using OpenMP, and ran on an Intel 8-core Xeon Processor. The processing speed is about 10 frames per second.
Fig. 6.
(a) An end-to-end trained small ConvNet and (b) LeNet with only the fully connected layer being trained while the rest (gray) of the layers inherited from the pretrained network trained on MNIST database.
2.7. Feature Region Measurement
CKD is defined as the progressive loss of kidney filtration function in time. Nephrons are the basic filtration unit in the kidney. A histology study of a rat CKD model has shown that when nephrons lose their filtration capability, the proximal convolutional tubules of those nephrons shrink and close.52 The rest of the functional nephrons in the kidney would compensate the dysfunctional ones. The proximal convolutional tubules of the functional nephrons enlarge and become hypertrophic. A histology study has shown that the average convolutional tubular diameter increases as CKD condition progresses.33
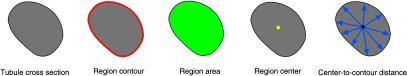
For each tubule’s cross-sectional region, the circularity of the region was first measured. Since a 2-D cross-sectional OCT image could cut the proximal tubule in any random plane, the circularity of the region indicates whether the tubule cross section is a perpendicular cut. Only the diameters of tubule regions with high circularity () are chosen for diameter analysis. The procedure to measure the circularity and diameter of a lumen is the following. (1) Locate the contour pixels of the region. The circumference of the region () is the total number of contour pixels. (2) Locate all the pixels that belong to the region. The area of the region () is the total number of pixels in the region. (3) Circularity of the region is calculated by dividing the circumference square of the region by times of region area, i.e., . A perfect circle would have a circularity equal to 1. (4) Locate the region center by averaging the coordinates of each pixel inside the region. (5) Distances from the region center to each contour pixel are measured. The diameter of the region is 2 times the mean of the distances. Figure 7 shows the process to determine the circularity and diameter of tubule cross-sectional region.
Fig. 7.
Illustration of definitions used in region measurements.
3. Results
3.1. Optical Coherence Tomography Imaging of Chronic Kidney Disease Rats
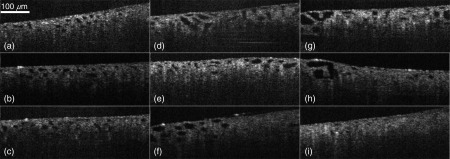
Visual differences between normal kidneys and CKD kidneys can be perceived from OCT images of rat kidneys. Images in Fig. 8 are collected from both healthy rats and rats with mid- to late-stage CKD conditions. In healthy kidneys, the proximal tubule lumens are similar in size and distributed uniformly with a homogeneous distribution pattern [Figs. 8(a)–8(c)]. In kidneys with late-stage ADR-induced CKD [Figs. 8(g)–8(i)], some proximal tubules either appear reduced in size or become indiscernible to OCT, i.e., atrophic. At the same time, the rest of the proximal tubules appear increased in size, i.e., hypertrophic. This is consistent with the published histology study in dysfunctional nephrons.33 These are the two most extreme cases in CKD kidney morphology. In the midstage CKD (week 2 to 6), the observed kidney images [Figs. 8(d)–8(f)] can be a mixture of these two extreme cases, where the hypertrophic tubules are smaller than those at late-stage and atrophic tubules are more discernible than those at late stage.
Fig. 8.
(a)–(c) are images captured from healthy kidney, where proximal tubules are uniform in size, (d)–(f) are images captured from midstage CKD kidney, where certain percentages of proximal tubules appear shrunk and certain percentages of proximal tubules appear enlarged, (g)–(i) are images captured from late-stage CKD kidney, where proximal tubules in some cross sections appear further enlarged and in some other cross sections disappeared due to shrunk to extreme small sizes.
3.2. Individual Image Inspection
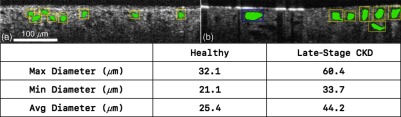
While it is possible to determine whether CKD exists, it is difficult to quantify the severity of the disease. To quantify the progression, we developed a CAD software to analyze the OCT images and measure the size of the proximal tubules. The CAD software was first tested on individual OCT images captured from different animals at different stages of CKD (Fig. 9). The software can detect and measure the tubular morphology and present the statistics instantly as the user navigates through images. In addition, the software highlights the detection using color-coded bounding boxes, where yellow, blue, and green indicate normal-size tubules, hypertrophic tubules (), and atrophic tubules (), respectively. Figure 9 shows two images examined by the software. Figures 9(a) and 9(b) are from a healthy kidney and a late-stage CKD (8 weeks after ADR injection) kidney, respectively. Visually, it is apparent that tubule lumens in Fig. 9(b) are larger than those from Fig. 9(a). The software measures the tubule lumen diameters, and the results from the software show that tubules from healthy kidneys are mostly within the normal ranges, which is 20 to . While the tubules from late-stage CKD kidneys are either in the hypertrophic range () or in the high end of normal ranges. The result is consistent with previous reports that the diameters of the hypertrophy tubules can be 50% larger in mice and nearly 100% larger in human than the diameters of the normal tubules.33,38 These results verify that the CAD software is able to detect and measure tubule cross sections accurately, which can be used to differentiate healthy and nonhealthy kidneys.
Fig. 9.
Individual image analysis shows significant tubular morphological differences between (a) healthy kidneys and (b) late-stage CKD kidneys.
3.3. Automatic Batch Analysis
In addition to individual image inspection, the software is able to analyze the entire batch of image dataset automatically. The processing speed is about 10 images per second. It takes about 10 min to process, measure, and generate statistics of a dataset containing 5000 images. The software records the information of each detected tubule, including diameter and circularity. The OCT image dataset can be quickly analyzed by the software without supervision of the user.
The complete OCT image dataset of rat kidneys was analyzed by the software. The dataset consists of 18 animals in total: 2 animals per week group for 8 post-ADR-injection weeks in total and 2 healthy animals for control. 3000 to 5000 2-D OCT images were collected per rat. In total, the complete dataset contains more than 70,000 images. The CAD software measures and records tubule diameters and generates the statistics, including tubular diameter, percentage of hypertrophic tubules, tubule density, and tubule diameter range for each week.
Tubule diameters are measured directly by the software after the tubule being detected. Percentage of hypertrophic tubules is derived by recording the number of detected tubules whose diameters are larger than . Tubule diameter range is the difference of the tubule diameter at 95 percentiles and that at 5 percentiles. Tubule density is computed by determining number of tubules per unit tissue area.
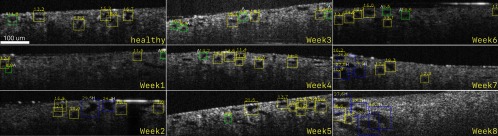
Figure 10 shows representative OCT images for the control and CKD-induced rat. For the healthy animal, most of the tubule diameters are within normal range (yellow). One week after injection, fewer tubules are visible and more atrophic tubules are present. During the midstages (week 2 to 6), number of both atrophic tubules and hypertrophic tubules increases. This is probably due to the fact that functional nephrons overwork to compensate the loss of atrophic tubules (green). At the late stages (week 7 to 8), more hypertrophic tubules (blue) are visible.
Fig. 10.
Sample kidney OCT images from healthy control rats and from CKD rats at each week after ADR injection. Bounding box color indicates the condition of the tubule, yellow color indicates normal tubule, green color (labeled with “A”) indicates atrophy tubule, and blue color (labeled with “H”) indicates hypertrophy tubule.
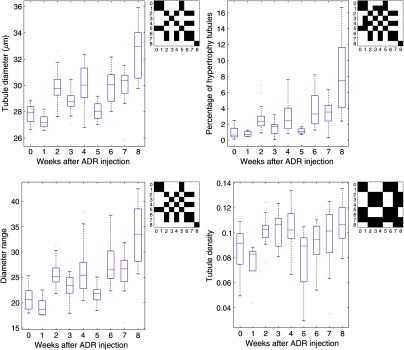
Figure 11 shows the statistical results from the analysis of the complete dataset. The statistics indicates a general trend of increase for average tubule diameter, diameter range, and percentage of hypertrophic tubules. The statistics also shows a fluctuation in these parameters during the midstages (week 2 to 6). We hypothesize it could be due to either variation of different rat’s reaction to ADR injection, sampling bias, or the recovery of the organ before late-stage CKD (week 7 to 8). Previous studies have observed renal function recovery in drug-induced acute kidney injury.53 Tubular density shows an initial increase (week 2 to 4) compared to normal, followed by a decrease in week 5 to 7. This could be explained by the closure of some atrophic tubules.
Fig. 11.
The trends of mean tubule diameter, percentage of hypertrophic tubules, tubule diameter range, and tubule density as CKD progression. Matrix of -test between each week is attached to the upper right corner of each plot. The dark regions indicate the -tests between two weeks produce value above 0.05 (insignificant), and the bright regions indicate -tests between two weeks produce value below 0.05 (significant).
4. Discussion
It is crucial that CKD is detected and treated at early stages.54 Currently, measuring glomerular filtration rate (GFR) score and observing the visual presence of kidney damage are the two commonly established CKD diagnosis methods in clinical nephrology.55
GFR score is a scoring system that describes kidney filtration capability. GFR score is calculated in terms of blood serum creatinine level test in addition to patient’s age, weight, and gender. Serum creatinine is a chemical waste produced by the body that is mainly filtered out by the kidneys. However, despite the wide use of GFR as the gold standard to estimate kidney function, studies have suggested that it is not sensitive to early-stage CKD. It has been shown that a significant change of serum creatinine concentration in blood does not occur until nearly 50% of the kidney is nonfunctional. It is because the rest of the functional nephrons can adapt and compensate the loss of filtration units.38
Examining the morphology of the kidney structures, including glomerulus and proximal convoluted tubules, using histology images is a more direct and accurate way to detect CKD. The correlation among nephron tubule morphology, nephron function, and CKD progression has been discussed extensively and been corroborated with histology images in literature.33,38,52,56–58 If a nephron loses its filtration capability due to injury or diseases, its tubules would appear reduced in size. Meanwhile, remaining functional nephrons become enlarged to compensate the loss of filtration capability.38 Although observing kidney morphology is an effective way to detect CKD in early stages, the current procedure requires a biopsy to acquire a slice of renal tissue for histology. Such procedure has many disadvantages. (1) It requires the removal of a certain amount of kidney tissues. Due to its invasiveness, only a small percentage of CKD patients receive diagnosis based on morphology images. (2) The histology sample size is restricted because of the limit amount of kidney tissue that can be sampled. The morphology observed with such technique is highly localized. Therefore, the observed condition may not accurately represent the condition of the entire organ. (3) It is a demanding and time-consuming task to acquire and prepare kidney biopsy samples. Such process includes delicate surgery procedure to acquire the sample, careful sample preservation with correct storage temperature and solutions, and sample staining for microscopy imaging. (4) Similar to many imaging-based diagnosis procedures, histology image assessment is a meticulous and time-consuming task that requires two experienced histopathologists to reduce the diagnosis errors.
OCT imaging can observe nephron tubular morphology without the need of acquiring tissue sample. This imaging method can examine the kidney structure in a similar fashion as histology reduces the invasiveness of such procedure. In addition, since it does not require tissues to be removed from the organ for observation, OCT imaging is able to image multiple locations in the kidney or even a full surface scan without altering the condition of the organ. Being able to image multiple locations in the kidney can provide a much more comprehensive view of the overall health condition of the organ compared to biopsy/histology, which uses a very small percentage of the organ tissue to assess the condition of the entire organ.
A detailed multilocation imaging of the kidney drastically increases number of images to inspect per patient for a more accurate diagnosis. Being able to process such large quantity of information on time has a significant impact on the diagnosis workflow. The presented CAD software showcases how to detect, measure, and statistically quantify kidney function based on OCT images. The software is not developed with the goal to replace the role of medical doctors; instead, it is developed as a useful tool for medical doctors to quickly scan through the large image database and compare their results with the statistics report generated by the software. The ultimate goal for such software is to decrease diagnosis time span, decrease diagnosis error rate, and improve the day-to-day workflow efficiency of medical doctors.
The current methods have limitations and room for improvement in several areas. (1) The animal experiment design to observe the progression of CKD can be improved. The current method can observe a single animal only once because it is very difficult to perform surgery on the same rat repeatedly for consecutive weeks. Therefore, the statistical results derived from this experiment design cannot avoid the biological variation between animals. Continuous monitoring of the kidney morphology of the same animal would definitely present a better picture of the morphology development with the CKD progression, especially at week 2 to 6. This will require an abdominal window.59 (2) The selection of imaging locations on the kidney can be biased by the operator. The bias induced by imaging location selection could also have an impact on the accuracy of the statistical results. The OCT instrument in use is a stationary microscope that performed scanning in a predefined rectangular region. Despite the fact that multiple locations (rectangular regions) have been scanned per each kidney, a scan covering the entire organ surface will provide the most unbiased description of the kidney condition. (3) The OCT probe needs to be held closely to the kidney surface to image the subsurface structures of the organ. To reduce the invasiveness of such procedure, laparoscope- or needle-based OCT probe would produce much smaller incision size and shorten the recovery period postsurgery.60
5. Conclusion
We demonstrated that OCT is able to evaluate living kidney microstructure and function in CKD models of Munich-Wistar rats. The preliminary results show that OCT can provide more information beyond proteinurea and serum creatinine. In addition, we presented a CAD software that can automatically detect, measure, and quantify features in the kidney images that are related to CKD progression. The software is necessary to process a large number of OCT images in a short period of time. Our ability of discern pathological changes in rat models of CKD is important in view of our recent studies showing that OCT can be used to evaluate the status of human kidneys in the operating room and its potential use in conjunction with renal biopsies to evaluate kidney histopathology in patients.
Acknowledgments
This work was supported by the National Institutes of Health under Grants Nos. R21AG042700, R01DK094877, and R21DK088066. We thank Professor James Fujimoto (Massachusetts Institute of Technology) for insightful discussion. We also thank Thomas W. Castonguay, Lily Jin, Zach Langley, and Phillip Liu for technical assistance.
Biographies
Bohan Wang received his BS degree in electrical and computer engineering from the University of Maryland, College Park, USA, in 2014. He is currently a doctoral candidate in electrical and computer engineering at the University of Maryland, College Park. His research interests include medical image analysis, computer vision, and machine learning.
Biographies for the other authors are not available.
Disclosures
The authors have no relevant financial interests in the paper and no other potential conflicts of interest to disclose.
References
- 1.Abboud H., Henrich W. L., “Stage IV chronic kidney disease,” N. Engl. J. Med. 362(1), 56–65 (2010). 10.1056/NEJMcp0906797 [DOI] [PubMed] [Google Scholar]
- 2.Nahas M. E. L., “The global challenge of chronic kidney disease,” Kidney Int. 68(6), 2918–2929 (2005). 10.1111/j.1523-1755.2005.00774.x [DOI] [PubMed] [Google Scholar]
- 3.Levey A. S., et al. , “National Kidney Foundation practice guidelines for chronic kidney disease: evaluation, classification, and stratification,” Ann. Intern. Med. 139(2), 137–147 (2003). 10.7326/0003-4819-139-2-200307150-00013 [DOI] [PubMed] [Google Scholar]
- 4.Stevens L. A., et al. , “Assessing kidney function—measured and estimated glomerular filtration rate,” N. Engl. J. Med. 354(23), 2473–2483 (2006). 10.1056/NEJMra054415 [DOI] [PubMed] [Google Scholar]
- 5.Huang D., et al. , “Optical coherence tomography,” Science 254(5035), 1178–1181 (1991). 10.1126/science.1957169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmitt J. M., “Optical coherence tomography (OCT): a review,” IEEE J. Sel. Top. Quantum Electron. 5(4), 1205–1215 (1999). 10.1109/2944.796348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reif R., Wang R. K., “Optical microangiography based on optical coherence tomography,” Chapter 45 in Optical Coherence Tomography, Drexler W., Fujimoto J. G., Eds., pp. 1373–1397, Springer International Publishing, Cham: (2015). [Google Scholar]
- 8.Chen Y., et al. , “High-resolution three-dimensional optical coherence tomography imaging of kidney microanatomy ex vivo,” J. Biomed. Opt. 12(3), 034008 (2007). 10.1117/1.2736421 [DOI] [PubMed] [Google Scholar]
- 9.Boppart S. A., et al. , “High-resolution optical coherence tomography-guided laser ablation of surgical tissue,” J. Surg. Res. 82(2), 275–284 (1999). 10.1006/jsre.1998.5555 [DOI] [PubMed] [Google Scholar]
- 10.Andrews P. M., et al. , “High-resolution optical coherence tomography imaging of the living kidney,” Lab. Invest. 88(4), 441–449 (2008). 10.1038/labinvest.2008.4 [DOI] [PubMed] [Google Scholar]
- 11.Andrews P. M., et al. , “Optical coherence tomography of the aging kidney,” Exp. Clin. Transplant. 14(6), 617–622 (2016). 10.6002/ect.2015.0212 [DOI] [PubMed] [Google Scholar]
- 12.Wierwille J., et al. , “In vivo, label-free, three-dimensional quantitative imaging of kidney microcirculation using Doppler optical coherence tomography,” Lab. Invest. 91(11), 1596–1604 (2011). 10.1038/labinvest.2011.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhi Z., et al. , “Highly sensitive imaging of renal microcirculation in vivo using ultrahigh sensitive optical microangiography,” Biomed. Opt. Express 2(5), 1059–1068 (2011). 10.1364/BOE.2.001059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andrews P. M., et al. , “Optical coherence tomography of the living human kidney,” J. Innovative Opt. Health Sci. 7(2), 1350064 (2014). 10.1142/S1793545813500648 [DOI] [Google Scholar]
- 15.Onozato M. L., et al. , “Optical coherence tomography of human kidney,” J. Urol. 183(5), 2090–2094 (2010). 10.1016/j.juro.2009.12.091 [DOI] [PubMed] [Google Scholar]
- 16.Cable A., et al. , “Automated quantification of microstructural dimensions of the human kidney using optical coherence tomography (OCT),” Opt. Express 17(18), 16000–16016 (2009). 10.1364/OE.17.016000 [DOI] [PubMed] [Google Scholar]
- 17.Andrews P. M., Chen Y., “Using optical coherence tomography (OCT) to evaluate human donor kidneys prior to and following transplantation,” J. Nephrol. Ther. 4, 151 (2014). 10.4172/2161-0959.1000151 [DOI] [Google Scholar]
- 18.Andrews P. M., et al. , “Mannitol infusion within 15 min of cross-clamp improves living donor kidney preservation,” Transplantation 98(8), 893–897 (2014). 10.1097/TP.0000000000000154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Templin C., et al. , “Vascular lesions induced by renal nerve ablation as assessed by optical coherence tomography: pre-and post-procedural comparison with the Simplicity® catheter system and the EnligHTN™ multi-electrode renal denervation catheter,” Eur. Heart J. 34(28), 2141–2148 (2013). 10.1093/eurheartj/eht141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barwari K., et al. , “Advanced diagnostics in renal mass using optical coherence tomography: a preliminary report,” J. Endourol. 25(2), 311–315 (2011). 10.1089/end.2010.0408 [DOI] [PubMed] [Google Scholar]
- 21.Barwari K., et al. , “Differentiation between normal renal tissue and renal tumours using functional optical coherence tomography: a phase I in vivo human study,” BJU Int. 110(8b), E421 (2012). 10.1111/j.1464-410X.2012.11220.x [DOI] [PubMed] [Google Scholar]
- 22.Doi K., “Current status and future potential of computer-aided diagnosis in medical imaging,” Br. J. Radiol. 78, S3–S19 (2014). 10.1259/bjr/82933343 [DOI] [PubMed] [Google Scholar]
- 23.Liu J., et al. , “Computer aided detection of epidural masses on computed tomography scans,” Comput. Med. Imaging Graphics 38(7), 606–612 (2014). 10.1016/j.compmedimag.2014.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farag A., et al. , “A bottom-up approach for automatic pancreas segmentation in abdominal CT scans,” Lect. Notes Comput. Sci. 8676, 103–113 (2014). 10.1007/978-3-319-13692-9 [DOI] [Google Scholar]
- 25.Jerebko A. K., et al. , “Multiple neural network classification scheme for detection of colonic polyps in CT colonography data sets,” Acad. Radiol. 10(2), 154–160 (2003). 10.1016/S1076-6332(03)80039-9 [DOI] [PubMed] [Google Scholar]
- 26.Wan S., et al. , “Integrated local binary pattern texture features for classification of breast tissue imaged by optical coherence microscopy,” Med. Image Anal. 38, 104–116 (2017). 10.1016/j.media.2017.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qi X., et al. , “Computer-aided diagnosis of dysplasia in Barrett’s esophagus using endoscopic optical coherence tomography,” J. Biomed. Opt. 11(4), 044010 (2006). 10.1117/1.2337314 [DOI] [PubMed] [Google Scholar]
- 28.Qi X., et al. , “Automated quantification of colonic crypt morphology using integrated microscopy and optical coherence tomography,” J. Biomed. Opt. 13(5), 054055 (2008). 10.1117/1.2993323 [DOI] [PubMed] [Google Scholar]
- 29.Roth H. R., et al. , “A new 2.5D representation for lymph node detection using random sets of deep convolutional neural network observations,” Lect. Notes Comput. Sci. 8673, 520–527 (2014). 10.1007/978-3-319-10404-1_65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roth H. R., et al. , “Deep convolutional networks for pancreas segmentation in CT imaging,” Proc. SPIE 9413, 94131G (2015). 10.1117/12.2081420 [DOI] [Google Scholar]
- 31.Liu J., et al. , “Colitis detection on abdominal CT scans by rich feature hierarchies,” Proc. SPIE 9785, 97851N (2016). 10.1117/12.2217681 [DOI] [Google Scholar]
- 32.Esteva A., et al. , “Dermatologist-level classification of skin cancer with deep neural networks,” Nature 542(7639), 115–118 (2017). 10.1038/nature21056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee V. W. S., et al. , “Adriamycin nephropathy in severe combined immunodeficient (SCID) mice,” Nephrol. Dial. Transplant. 21, 3293–3298 (2006). 10.1093/ndt/gfl413 [DOI] [PubMed] [Google Scholar]
- 34.Li Z., et al. , “Monitoring kidney microanatomy changes during ischemia-reperfusion process using texture analysis of OCT images,” IEEE Photonics J. 9(2), 1–10 (2017). 10.1109/JPHOT.2017.2669482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang Q., et al. , “Depth-resolved imaging of colon tumor using optical coherence tomography and fluorescence laminar optical tomography,” Biomed. Opt. Express 7(12), 5218–5232 (2016). 10.1364/BOE.7.005218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ding Z., et al. , “Imaging spinal structures with polarization-sensitive optical coherence tomography,” IEEE Photonics J. 8(5), 1–8 (2016). 10.1109/JPHOT.2016.2605447 [DOI] [Google Scholar]
- 37.Tang Q., et al. , “Real-time epidural anesthesia guidance using optical coherence tomography needle probe,” in CLEO: Applications and Technology, p. AM2O.3, OSA, Washington DC: (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chevalier R. L., “The proximal tubule is the primary target of injury and progression of kidney disease: role of the glomerulotubular junction,” Am. J. Physiol. 311(1), F145–F161 (2016). 10.1152/ajprenal.00164.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Viola P., Jones M. J., “Robust real-time face detection,” Int. J. Comput. Vision 57(2), 137–154 (2004). 10.1023/B:VISI.0000013087.49260.fb [DOI] [Google Scholar]
- 40.Friedman J. H., “On bias, variance, 0/1—loss, and the curse-of-dimensionality,” Data Min. Knowl. Discovery 1(1), 55–77 (1997). 10.1023/A:1009778005914 [DOI] [Google Scholar]
- 41.Land E. H., McCann J. J., “Lightness and retinex theory,” J. Opt. Soc. Am. 61(1), 1–11 (1971). 10.1364/JOSA.61.000001 [DOI] [PubMed] [Google Scholar]
- 42.Crow F. C., “Summed-area tables for texture mapping,” ACM SIGGRAPH Comput. Graphics 18(3), 207–212 (1984). 10.1145/964965 [DOI] [Google Scholar]
- 43.Bradley D., Roth G., “Adaptive thresholding using the integral image,” J. Graphics GPU Game Tools 12(2), 13–21 (2011). 10.1080/2151237X.2007.10129236 [DOI] [Google Scholar]
- 44.Felzenszwalb P. F., Huttenlocher D. P., “Efficient graph-based image segmentation,” Int. J. Comput. Vision 59(2), 167–181 (2004). 10.1023/B:VISI.0000022288.19776.77 [DOI] [Google Scholar]
- 45.Levinshtein A., et al. , “Turbopixels: fast superpixels using geometric flows,” IEEE Trans. Pattern Anal. Mach. Intell. 31(12), 2290–2297 (2009). 10.1109/TPAMI.2009.96 [DOI] [PubMed] [Google Scholar]
- 46.Achanta R., et al. , “SLIC superpixels compared to state-of-the-art superpixel methods,” IEEE Trans. Pattern Anal. Mach. Intell. 34(11), 2274–2282 (2012). 10.1109/TPAMI.2012.120 [DOI] [PubMed] [Google Scholar]
- 47.Turk M. A., Pentland A. P., “Face recognition using eigenfaces,” in Int. Conf. on Computer Vision and Pattern Recognition, pp. 586–591, IEEE; (1991). 10.1109/CVPR.1991.139758 [DOI] [Google Scholar]
- 48.LeCun Y., et al. , “Gradient-based learning applied to document recognition,” Proc. IEEE 86(11), 2278–2324 (1998). 10.1109/5.726791 [DOI] [Google Scholar]
- 49.Krizhevsky A., Sutskever I., Hinton G. E., “ImageNet classification with deep convolutional neural networks,” in Neural Information Processing Systems, pp. 1097–1105 (2012). [Google Scholar]
- 50.Yosinski J., Clune J., Bengio Y., “How transferable are features in deep neural networks?” in Advances in Neural Information Processing Systems, pp. 3320–3328 (2014). [Google Scholar]
- 51.Vedaldi A., Lenc K., “MatConvNet: convolutional neural networks for MATLAB,” in MM ’15 Proc. of the 23rd ACM Int. conf. on Multimedia, pp. 689–692, ACM, New York, New York: (2015). 10.1145/2733373.2807412 [DOI] [Google Scholar]
- 52.Wang Y., et al. , “Progressive adriamycin nephropathy in mice: sequence of histologic and immunohistochemical events,” Kidney Int. 58, 1797–1804 (2000). 10.1046/j.1523-1755.2000.00342.x [DOI] [PubMed] [Google Scholar]
- 53.Singh N. P., Ganguli A., Prakash A., “Drug-induced kidney diseases,” J. Assoc. Physicians India 51, 970–979 (2003). [PubMed] [Google Scholar]
- 54.El Nahas A. M., Bello A. K., “Chronic kidney disease: the global challenge,” Lancet 365(9456), 331–340 (2005). 10.1016/S0140-6736(05)17789-7 [DOI] [PubMed] [Google Scholar]
- 55.Levey A. S., Coresh J., “Chronic kidney disease,” Lancet 379(9811), 165–180 (2012). 10.1016/S0140-6736(11)60178-5 [DOI] [PubMed] [Google Scholar]
- 56.Lee V. W., Harris D. C., “Adriamycin nephropathy: a model of focal segmental glomerulosclerosis,” Nephrology 16(1), 30–38 (2011). 10.1111/nep.2011.16.issue-1 [DOI] [PubMed] [Google Scholar]
- 57.Metcalfe W., “How does early chronic kidney disease progress? A background paper prepared for the UK Consensus Conference on early chronic kidney disease,” Nephrol. Dial. Transplant. 22(Suppl. 9), ix26–ix30 (2007). 10.1093/ndt/gfm446 [DOI] [PubMed] [Google Scholar]
- 58.Evan A. P., et al. , “Proximal tubule morphology after single nephron obstruction in the rat kidney,” Kidney Int. 30(6), 818–827 (1986). 10.1038/ki.1986.261 [DOI] [PubMed] [Google Scholar]
- 59.Ritsma L., et al. , “Surgical implantation of an abdominal imaging window for intravital microscopy,” Nat. Protoc. 8(3), 583–594 (2013). 10.1038/nprot.2013.026 [DOI] [PubMed] [Google Scholar]
- 60.Tan H.-J., et al. , “Population level assessment of hospital based outcomes following laparoscopic versus open partial nephrectomy during the adoption of minimally invasive surgery,” J. Urol. 191(5), 1231–1237 (2014). 10.1016/j.juro.2013.11.002 [DOI] [PubMed] [Google Scholar]