Fig. 7.
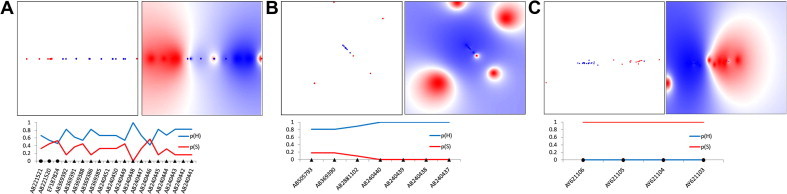
Host-specific separation of HEV3 and HEV4 strains in LP-modeled physicochemical space. Shown are LP plots of physicochemical properties for aa sites from Pol and Pp (Table 4). Probability mapping of human and swine strains is color-coded, with human space shown in blue and swine in red. Color density is proportional to probability values. (A) LP map of HEV3 variants (n = 65) using Pol aa physicochemical properties or markers (n = 16); (B) LP map of HEV4 variants (n = 55) using Pol markers (n = 16) and (C) LP map of HEV4 variants (n = 55) using the Pp markers (n = 10). Below the mappings are line charts showing the prediction results (probability scores) on validation datasets from the above corresponding LP maps; y-axis represents probability [0–1]; p(H) and p(S) are probabilities of the human (blue line) and swine (red line) origin of a strain, respectively. GenBank accession numbers (x-axis) are shown for each test sequence; black triangles and circles identify HEV strains obtained from humans and swine, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
