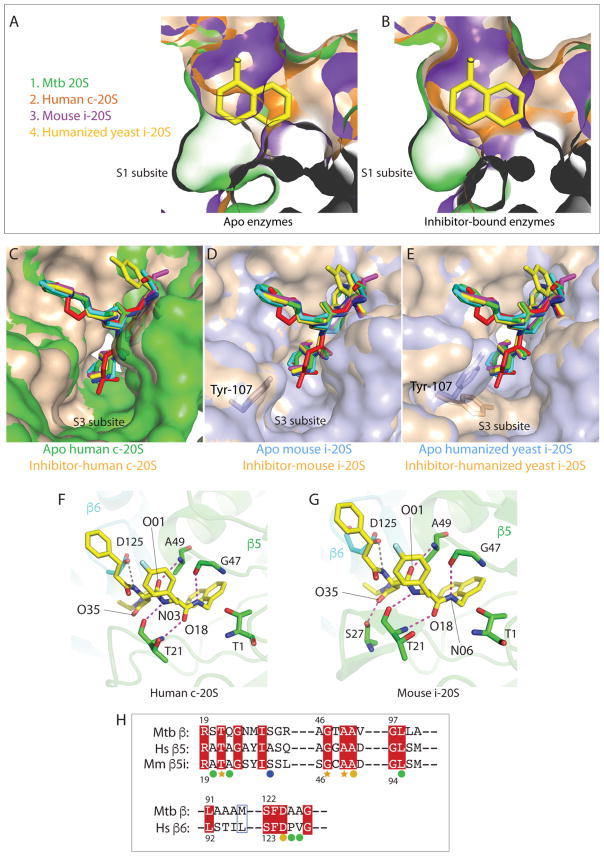
Figure 5. Proposed interactions between dipeptide inhibitors and the chymotrypsin-like active site of c-20S and i-20S.
.(A) The apo-Mtb20SOG (PDB ID 3HFA) structure is aligned with ligand-free human c-20S β5/β6 (PDB ID 4R3O), mouse i-20S β5i/β6 (PDB ID 3UNH), and humanized yeast i-20S (PDB ID 5L5B). The S1 binding pockets of c-20S and i-20S cannot accommodate the P1 naphthyl group without a conformational change. Human β5/β6 is in orange; mouse β5i/β6 is in purple; apo-Mtb20SOG is in green; humanized yeast β5i/β6 is in wheat. (B) DPLG-2-bound Mtb20SOG is modeled into carfilzomib-bound human β5/β6 (PDB ID 4R67), PR-957-bound mouse β5i/β6 (PDB ID 3UNF) and carfilzomib-bound humanized yeast β5i/β6 (PDB ID 5L5E). After conformational changes, the S1 pockets of β5/β6 and β5i/β6 are enlarged to accommodate the naphthyl group. (C) The P3 groups of the six dipeptides are modeled into the S3 pocket of the human c-20S. Both ligand-free and ligand-bound S3 pockets have enough room to accommodate various P3 groups. The apo-β5/β6 is in green and ligand-bound β5/β6 is in wheat. (D) The P3 groups of the six inhibitors are modeled into the S3 pockets of mouse i-20S. No further conformational change is required to fit the P3 groups after ligand binding. The apo- β5i/β6 is in light blue and ligand-bound β5i/β6 is in wheat. (E) The P3 groups of the six inhibitors are modeled into the S3 pockets of the humanized yeast β5i/β6. Tyr107 of β6 rotates to widen the entrance of the S3 pocket, likely facilitating inhibitor binding. The humanized yeast apo-β5i/β6 is in light blue and ligand-bound β5i/β6 is in wheat. (F) Proposed hydrogen bonding interactions between DPLG-2 and the human c-20S β5/β6. The antiparallel bindings between inhibitor backbone and Thr-21, Gly-47, and Ala-49 are conserved in Mtb 20S and in human c-20S and mouse i-20S. The β5 subunit is in green and β6 is in cyan. The grey dashed lines are the potential hydrogen bonds within 3.6 Å distance. (G) Modeled interactions of DPLG-2 with mouse i-20S β5i/β6. Besides the backbone interactions, Ser-27 contributes additional H-bond to the O35 of the P3 group. (H) Upper panel: Sequence alignment of Mtb 20S β subunit, human c-20S β5 subunit, and mouse i-20S β5i subunit. Lower panel: Sequence alignment of Mtb 20S β subunit and human 20S β6 subunit. The orange stars denote the residues involved in backbone antiparallel bindings that are conserved in all three proteasomes. The orange circles indicate that the conserved residues participate in inhibitor binding in the Mtb proteasome and possibly in c-20S and i-20S. Green circles are residues specific for Mtb 20S involved in dipeptide inhibitor binding. Ser-27 (blue circle), which is not conserved in c-20S β5, is essential for P3 binding.