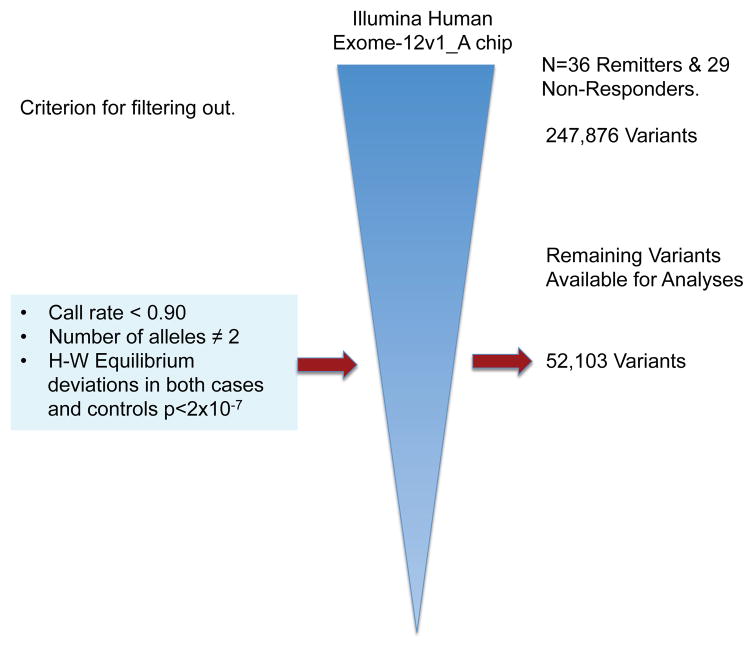
Fig 1. Process used to filter out SNP Exomic Variants Genotyped with the Illumina® HumanExome BeadChip-12v1_A in a Mexican-American Cohort of Patients with Major Depressive Disorder (MDD) under Antidepressive Treatment to Evaluate Pharmacogenetic Response As Remitters (n=36) and Non-Responders (n=29).
From 247,876 SNPs 195,773 were discarded because they were either monoallelic, had more than two alleles, had a call rate lower than 90%, or their genotype proportions deviated from the expected ones as defined by the Hardy-Weinberg equilibrium theorem in both cases (Remitters) and controls (Non-responders), P<2 × 10−7. The remaining 52,103 SNPs were used for Exome-Wide Association Analysis.