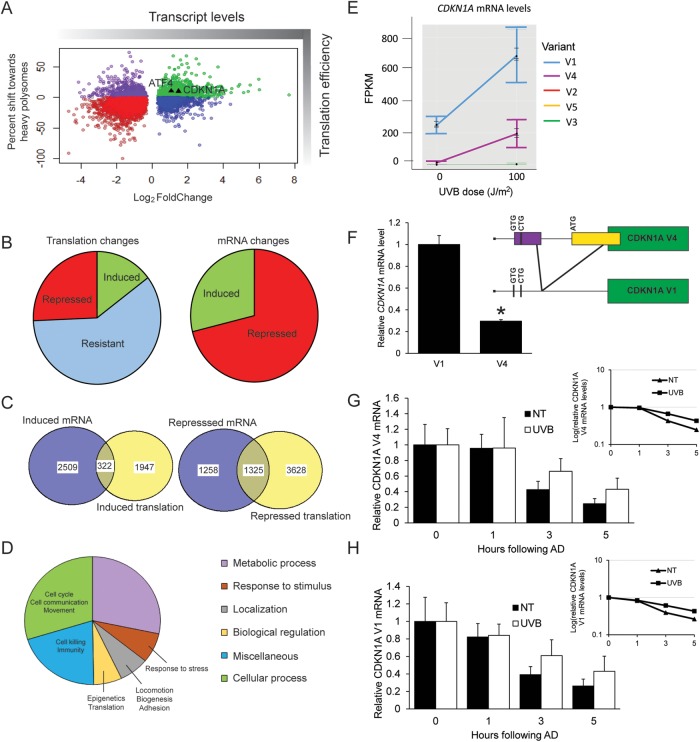
FIGURE 3:
Genome-wide analyses reveal preferential translation of CDKN1A following UVB. N-TERT keratinocytes were irradiated with 0 or 100 J/m2 UVB, followed by lysate preparation and polysome profiling. RNA was isolated from total cell lysate and light or heavy polysome fractions, and each was analyzed by RNA-Seq. (A) Scatterplot illustrating the shift toward heavy polysomes vs. relative mRNA fold changes following UVB irradiation. Only genes with significant mRNA fold changes (p < 0.05) are shown. (B) On the left side of the panel is a pie chart indicating the numbers of genes whose translation was induced (>10% shift), resistant (between -10% and 10% shift), or repressed (less than -10% shift) following UVB irradiation. On the right side of the panel is the number of genes whose transcripts were significantly induced or repressed after UVB treatment. (C) Venn diagram comparing the gene transcripts with significantly induced mRNA following UVB treatment and those genes that are subject to induced translation, as judged by a >10% shift of the encoded mRNAs toward large polysomes. Additionally, a Venn diagram is featured showing those gene transcripts that were significantly repressed following UVB irradiation, along with those genes displaying translation repression (>10% shift toward small polysomes). (D) Pie chart illustrating the different cellular functions of genes that are suggested to be preferentially translated in response to UVB. (E) FPKM values to indicate relative expression level for each CDKN1A transcript variant after the indicated UVB dose. (F) Pattern of alternative splicing of the CDKN1A, leading to V1 and V4 transcripts that display the uORFs and coding sequences (green boxes). Potential initiation codons are illustrated in the 5′-leader of the V1 transcript. Primers that recognize both V1 and V4 were used to perform PCR on N-TERT keratinocytes to determine relative transcript abundance. Bands were quantified by densitometry and normalized to PCR product size. N-TERTs were exposed to 0 or 100 J/m2 UVB, and after 8 h, RNA was prepared from the cells for measurement of V4 (G) and V1 (H) mRNAs by qPCR. Additionally, cells were treated with 10 μM actinomycin D (AD) and cultured for an additional 1, 3, or 5 h, and levels of the V1 and V4 mRNAs were determined to assess the rate of decay for these transcripts. Transcript levels were measured in no treatment (NT) or UVB exposed cells following actinomycin treatments were normalized to the respective cells measured in the absence of actinomycin D treatment (0). A semilog plot of each decay rate is shown in the inset plot for each transcript. Error bars represent mean ± SD of three biological replicates. * indicates p < 0.05.