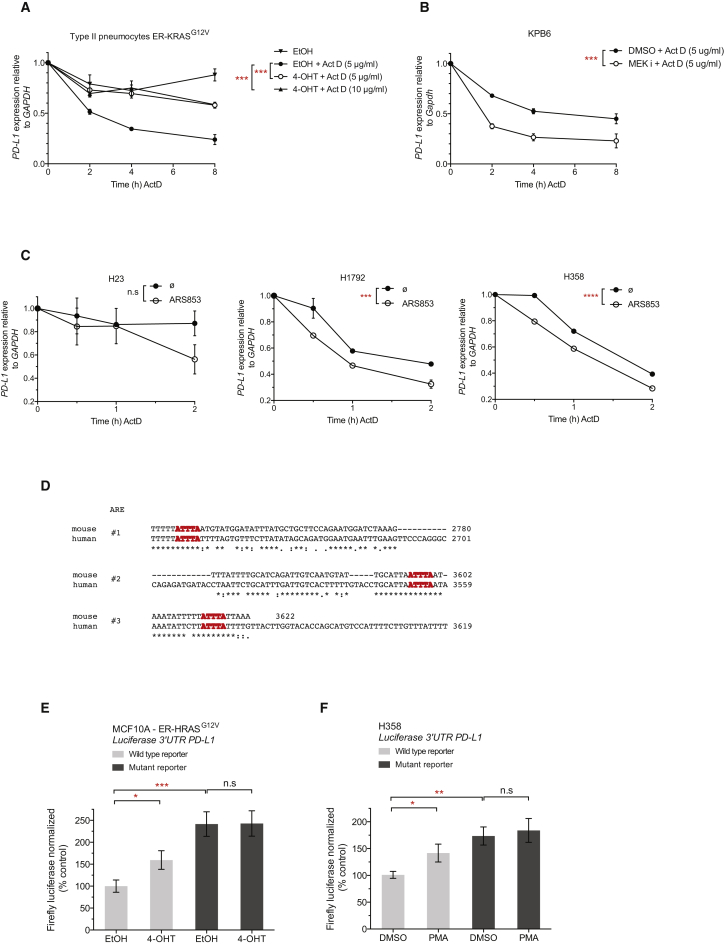
Figure 2.
RAS Signaling Increases PD-L1 mRNA Stability through AU-Rich Elements in the 3′ UTR
(A) qPCR analysis of PD-L1 mRNA stability in ER-KRASG12V type II pneumocytes after the concomitant addition of actinomycin D (5 μg/mL or 10 μg/mL) and 4-OHT or vehicle added at time = 0 hr in starvation medium. Mean ± SEM of two independent experiments. ∗∗∗p < 0.0005; two-way ANOVA.
(B) qPCR analysis of PD-L1 mRNA stability in KPB6 cells after the addition of actinomycin D (5 μg/mL) and DMSO or MEK inhibitor. Cells were pre-treated with DMSO or MEK inhibitor for 30 min before actinomycin D addition. Mean ± SEM of two independent experiments. ∗∗∗p < 0.0005; two-way ANOVA.
(C) qPCR analysis of PD-L1 mRNA stability after the addition of actinomycin D (5 μg/mL) and DMSO or ARS853. Cells were pre-treated with DMSO or ARS853 for 35 min before actinomycin D addition. Mean ± SEM of two independent experiments. ∗∗∗p < 0.0005; two-way ANOVA.
(D) Sequence alignment of conserved AU-rich element ATTTA pentamer sequences (highlighted in red) in the mouse and human CD274 3′ UTR.
(E) Normalized luciferase signal in ER-HRASG12V MCF10A cells from wild-type (ATTTA x 6) or mutant (ATGTA x 6) PD-L1 3′ UTR reporters, 24 hr after treatment in starvation medium. Mean ± SEM of three independent experiments.
(F) Normalized luciferase signal in H358 cells from wild-type (ATTTA x 6) or mutant (ATGTA x 6) PD-L1 3′ UTR reporters, 6 hr after treatment. Mean ± SEM of three independent experiments.
Abbreviations and quantities: 4-OHT, 100 nM; MEK inhibitor GSK1120212, 25 nM; PMA, 200 nM. ∗∗∗p < 0.0005, ∗∗p < 0.005, ∗p < 0.05, n.s., not significant. Unpaired, two-tailed Student’s t tests. See also Figure S2.