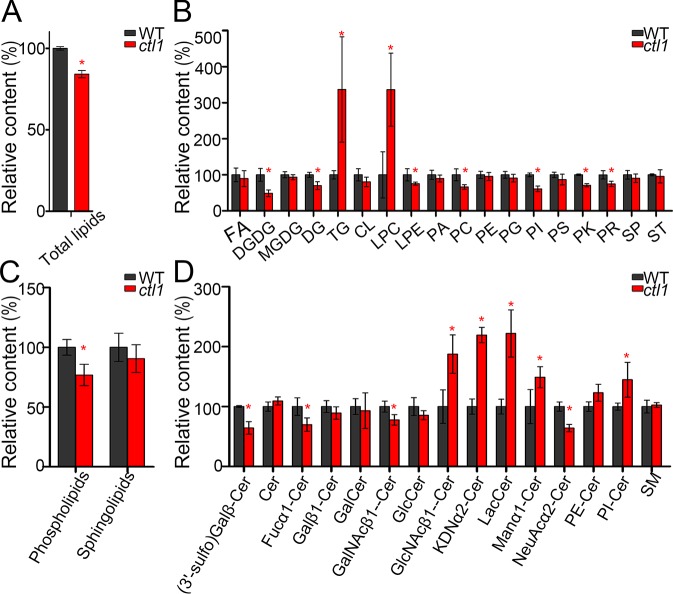
Fig 8. The lipidomics analysis of wild type and ctl1.
(A) The relative content of total lipids in the wild type (WT) and the ctl1 mutant. (B) The changes in the subclasses of lipids in the ctl1 mutant. Fatty acyls (FAs), digalactosyldiacylglycerol (DGMG), monogalactosyldiacylglycerol (MGDG), diacylglycerol (DG), triacylglycerol (TG), lysophosphatidylcholine (LPC), lysophosphatidylethanolamine (LPE), phosphatidic acid (PA), phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylglycerol (PG), phosphatidylinositol (PI), phosphatidylserine (PS), polyketides (PKs), prenol lipids (PRs), sphingolipids (SPs), and sterol lipids (STs). (C) The relative intensity of total glycerophospholipids and SPs in the WT and the ctl1 mutant. (D) Some subclasses of SPs are significantly altered in the ctl1 mutant. These include a group of ceramides (Cer) such as Fucα1-Cer: Fucα1-2GalNAcβ1-4(NeuAcα2-8NeuAcα2–3) Galβ1-4Glcβ-Cer; Galβ1-Cer: Galβ1-3GalNAcβ1-3Galα1-3Galβ1-4Glcβ-Cer; GalNAcβ1-Cer: GalNAcβ1-3Galα1-3Galα1-3Galα1-3Galα1-4Galβ1-4Glcβ-Cer; GlcNAcβ1-Cer: GlcNAcβ1-4Manβ1-4Glcβ-Cer; KDNα2-Cer: KDNα2-6Galβ1-4GlcNAcβ1-3Galβ1-4Glcβ-Cer; Manα1-Cer: Manα1-3Manβ1-4Glcβ-Cer; and NeuAcα2-Cer: NeuAcα2-8NeuAcα2-3Galβ1-4Glcβ-Cer. The classification of these SPs was from the method provided by the website LIPID Metabolites and Pathways Strategy(LIPID MAPS) (http://www.lipidmaps.org/). Data are mean ± SD. Four independent experiments were performed. Red asterisks indicate a significant difference (*P < 0.05, Student t test). The raw data for panels A–D can be found in S1 Data.