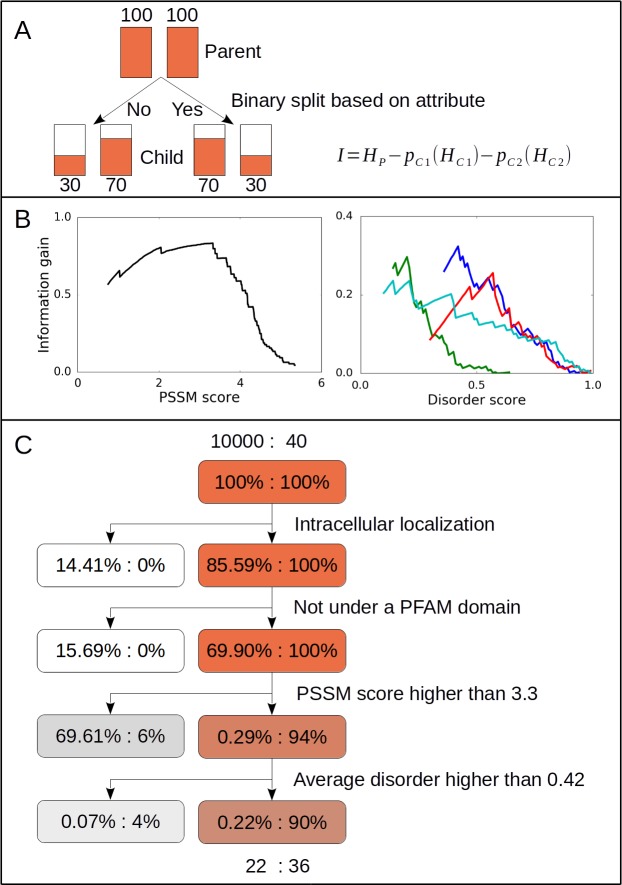
Fig 2. Filtering protocol to find true binding partners.
(A) Schematic diagram of the binary filtering protocol we created utilizing information gain. A given attribute provides a binary split of the Parent group into the C1 and C2 child groups. The information gain (I) is then calculated as the difference of the Shannon entropy (H) of the Parent group minus the Shannon entropy of the Children groups weighted by their relative probabilities (p). These values were calculated over the dataset containing 40 known human binding partners and 10,000 random human segments from the proteome with a higher than zero PSSM score. (B) The information gain of the PSSM score (left panel) and four disorder prediction methods as a function of different cut-off values (right panel). The disorder prediction method used here were: IUPred (blue), Espritz Disprot (green) and VSL2 (red line), DISOPRED3 (cyan). Optimal cut-off values were obtained from the cut-off value corresponding to the maximum of the information gain, yielding 3.3 for the PSSM score, and 0.42 for IUPred disorder prediction score. (C) The outline of the final filtering protocol indicating the number of elements and percentage of cases in each Child group with the applied binary split.