Abstract
Long non-coding RNAs (lncRNAs) have been demonstrated to regulate metabolic tissue development and function, including adipogenesis, hepatic lipid metabolism, islet function and energy balance. However, the role of lncRNAs in gluconeogenesis remains completely unknown. Metformin reduces glucose output mainly via the inhibition of gluconeogenesis. In the present study, the lncRNA expression profile of primary mouse hepatocytes exposed to cyclic adenosine monophosphate (cAMP), a gluconeogenic stimulus, with or without metformin was analyzed by microarray. Among the 22,016 lncRNAs that were identified, 456 were upregulated and 409 were downregulated by cAMP (fold-change ≥2.0). Furthermore, the cAMP-induced upregulation of 189 lncRNAs and downregulation of 167 lncRNAs was attenuated by metformin. The expression levels of eight lncRNAs were validated by reverse transcription-quantitative polymerase chain reaction, and the results were consistent with those of the microarray analysis. Among them, two lncRNAs NR_027710 and ENSMUST00000138573, were identified to have an association with two protein coding genes, namely peroxisome proliferator-activated receptor-γ coactivator-1α, a critical transcriptional coactivator in gluconeogenesis, and G protein-coupled receptor 155, respectively. The two protein coding genes exhibited similar expression patterns to their associated lncRNAs. The findings of the present study suggest that lncRNAs are potentially involved in the regulation of gluconeogenesis.
Keywords: long non-coding RNAs, gluconeogenesis, microarray, metformin, peroxisome proliferator-activated receptor-γ coactivator-1α
Introduction
The incidence of diabetes mellitus is increasing, and this condition is becoming one of the major causes of morbidity and mortality across the globe (1). Blood glucose levels are maintained within a narrow range in healthy individuals by the liver through the opposing actions of insulin and glucagon (2). Hepatic gluconeogenesis, which is inhibited by insulin and stimulated by glucagon, serves an essential role in the maintenance of a normal blood glucose level during fasting. Gluconeogenesis becomes unrestrained in diabetes due to either deficient insulin secretion in type 1 diabetes mellitus or deficient insulin action in type 2 diabetes mellitus, which contributes to hyperglycemia (3). Therefore, elucidation of the molecular mechanism involved in the regulation of hepatic gluconeogenesis is likely to provide new avenues for the treatment of type 2 diabetes mellitus.
Long non-coding RNAs (lncRNAs) are defined as transcripts of >200 nucleotides (nt) that do not encode protein. According to their position relative to nearby coding genes, lncRNAs can generally be classified into four categories, namely sense overlap, antisense overlap, bidirectional and intergenic noncoding RNAs (4). It has been suggested that lncRNAs serve a pivotal role in physiology and disease (5,6). Morán et al (7) uncovered hundreds of islet lncRNAs by strand-specific analysis, some of which were dysregulated in type 2 diabetes or mapped to genetic loci underlying diabetes susceptibility. In adipose tissue, numerous lncRNAs have been identified to regulate adipogenesis (8). In muscle cells, H19 LncRNA has been indicated to mediate the regulation of glucose metabolism (9). However, little is known about the role of lncRNAs in hepatic gluconeogenesis.
In the fasting state, the increased secretion of the catabolic hormone glucagon stimulates gluconeogenesis by triggering the cyclic adenosine monophosphate (cAMP)/protein kinase A pathway and promoting the transcription of gluconeogenic genes (10). Metformin is currently the first drug of choice for the treatment of type 2 diabetes mellitus (11). It has been demonstrated that metformin reduces glucose output mainly via the inhibition of gluconeogenesis (12). However, the exact mechanism remains unclear (13–15). To identify whether lncRNAs are involved in the metformin-mediated inhibition of gluconeogenesis, a systematic analysis of the lncRNA expression profile in cAMP-stimulated primary mouse hepatocytes was performed in the present study. The cAMP-induced changes in lncRNA expression that were attenuated by metformin were identified. Among them, the expression levels of eight lncRNAs were validated by reverse transcription-quantitative polymerase chain reaction (RT-qPCR). The aim of the present study was to identify the potential role of lncRNAs in the regulation of gluconeogenesis.
Materials and methods
Materials
A total of 48 C57Bl/6 mice (age, 8–12 weeks old; weight, 18–20 g) were purchased from Shanghai Slack Experimental Center (Shanghai, China) and were housed in a specific pathogen free (SPF) environment (24–26°C; relative humidity 50–60%) with a 12-h light/dark cycle and free access to food and water. Dulbecco's modified Eagle's medium (DMEM) and Hank's balanced salt solution (HBSS) were obtained from Gibco (Thermo Fisher Scientific, Inc., Waltham, MA, USA). Hepatocyte medium was purchased from ScienCell Research Laboratories, Inc. (Carlsbad, CA, USA). Sodium pyruvate, sodium lactate, dexamethasone, bovine serum albumin (BSA), 8-bromoadenosine 3′,5′-cyclic monophosphate (8-br-cAMP) and metformin were acquired from Sigma-Aldrich (Merck KGaA, Darmstadt, Germany). All the primers used in RT-qPCR were synthesized by Shanghai Sangon Biological Engineering Technology and Services Co., Ltd. (Shanghai, China).
Primary mouse hepatocyte isolation and culture
All experiments were supervised and approved by the laboratory ethics committee of Ruijin Hospital affiliated with Shanghai Jiaotong University School of Medicine (Shanghai, China). Hepatocytes were isolated from 8–12 week old male C57Bl/6 mice. Briefly, following anesthesia, mouse livers were perfused with 10 ml 1X HBSS without calcium, followed by perfusion with 0.05% collagenase IV in calcium-containing HBSS in a recirculating manner. The liver was then detached and filtered through a 70 μm nylon mesh and hepatocytes were sedimented by centrifugation at 200 × g for 5 min at 4°C. The hepatocytes were plated onto 6-well plates and grown in hepatocyte medium supplemented with BSA, penicillin/streptomycin and hepatocyte growth factor. The cultures were maintained at 37°C in humidified atmosphere (95% air and 5% CO2). After 24 h, the cells had become attached to the plates and the medium was replaced with low glucose DMEM containing 0.25% BSA with dexamethasone (100 nM) for prestimulation for 12–16 h. The cells were then incubated with 100 μM 8-br-cAMP (a permeable analogue of cAMP) in the presence or absence of 2 mM metformin in glucose-free DMEM containing gluconeogenic substrates (10 mM sodium lactate and 1 mM sodium pyruvate) for 8 h.
RNA extraction and quality control
Total RNA was extracted from primary mice hepatocytes using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The RNA purity and concentration was evaluated with a NanoDrop-2000 spectrophotometer (Thermo Fisher Scientific, Inc.). The optical density (OD) absorbance ratio at 260 and 280 nm (A260/A280 ratio) was ~2.0 and the OD A260/A230 ratio was ~1.8. RNA integrity was determined by standard denaturing agarose gel electrophoresis, which revealed that the samples were good quality.
Microarray analysis
ArrayStar mouse LncRNA microarray version 2.0 (ArrayStar Inc., Rockville, MD, USA) is designed for the global profiling of mouse lncRNAs in hepatocytes isolated from 3 mice (i.e., 3 groups at 3 replicates). The sample preparations and microarray hybridization were performed based on the manufacturer's protocols. Briefly, 1 μg total RNA from each sample was amplified and transcribed into fluorescent cRNA using Agilent's Quick Amp Labeling kits (Agilent Technologies, Inc., Santa Clara, CA, USA) according to the manufacturer's protocol. The labeled cRNAs were hybridized onto the mouse 8×60K LncRNA array. After washing, the arrays were scanned using the Agilent G2505C Microarray Scanner System, and Agilent Feature Extraction software was used to analyze the acquired array images (Agilent Technologies, Inc.). Quantile normalization and subsequent data processing were performed using the GeneSpring GX v12.0 software package (Agilent Technologies, Inc.). The threshold set for significantly differential genes was an absolute log value of fold-change ≥2 and a P-value ≤0.05. The microarray analysis was performed by KangChen Bio-tech Inc. Corporations (Shanghai, China).
RT-qPCR
Following total RNA extraction, as described above, cDNA was synthesized by reverse transcription using Superscript II RT reagent kit (Promega Corporation, Madison, WI, USA) according to the manufacturer's protocol. RT-qPCR was performed with the Roche LightCycler® 480 instrument (Roche Diagnostics, Basel, Switzerland) using SYBR Premix Ex Taq (Takara Bio, Inc., Otsu, Japan) in a final volume of 10 μl. The PCR conditions were as follows: Denaturation at 95°C for 10 sec, 40 cycles at 95°C for 5 sec and 60°C for 31 sec. A melting curve was constructed in the temperature range of 60 to 95°C at the end of the amplification. The sequences of the primers used are shown in Table I. Relative gene expression levels were quantified based on the cycle threshold (Cq) values and normalized to the internal control gene β-actin, which received a value of 1. The relative gene expression was calculated using the 2−ΔΔCq method (16).
Table I.
Sequence of the primers used for reverse transcription-quantitative polymerase chain reaction.
| Gene | Forward sequence (5′ to 3′) | Reverse sequence (5′ to 3′) |
|---|---|---|
| PEPCK | GTGCTGGAGTGGATGTTCGG | CTGGCTGATTCTCTGTTTCAGG |
| AK133602 | GACCAGTCTGACCCCTATCC | TAACCAGCCATTCACCCTC |
| ENSMUST00000138573 | CCTCAGCACAGAAACCACC | AGCCTGTCCAACTATGCGT |
| ENSMUST00000129953 | TTCAGCCTACAATCCCAAG | GTGGAATACACCCCGAAGT |
| uc009njr.1 | TTTGTCGCTCATCCACGCT | GCAAATGGTCCCGCAAATA |
| AA591058 | GACCACCAGGCAGCACTTC | ACCACACACACCCACACACAC |
| NR_027710 | TGCCATATCTTCCAGTGACC | TAAGGTTCGCTCAATAGTCTTG |
| NR_030715 | TGGGGTGGAGAAGACAACA | AGCACAACCACAAGCATCTAC |
| ENSMUST00000145208 | TCTCAACTCTATGCCGCCTT | GCACACTTCCGCTTCACCT |
| Gpr155 | GTTTTTCTGTGCCGTGTTTAACT | GGGGGATTCTCTGTCTGCT |
| PGC-1α | ATACCGCAAAGAGCACGAGAAG | CTCAAGAGCAGCGAAAGCGTCACAG |
| β-actin | GGCTGTATTCCCCTCCATCG | CCAGTTGGTAACAATGCCATGT |
PEPCK, phosphoenolpyruvate carboxykinase; Gpr155, G protein-coupled receptor 155; PGC-1α, peroxisome proliferator-activated receptor-γ coactivator-1α.
Statistical analysis
All values are presented as mean ± standard deviation from at least three independent experiments, and P≤0.05 was considered to indicate a statistically significant difference. Differential expression levels of genes were compared using ANOVA for multiple groups or Student's t-test for two groups. Statistical analysis was performed by SPSS 18.0 software (IBM, Armonk, NY, USA).
Results
Phosphoenolpyruvate carboxykinase (PEPCK) expression levels under different conditions
Gluconeogenesis is tightly controlled through the transcriptional regulation of PEPCK, the rate-limiting enzyme of hepatic gluconeogenesis (17). Metformin has been reported to suppress PEPCK expression in primary hepatocytes (14). Prior to microarray analysis, the effect of metformin on PEPCK mRNA expression in the primary mouse hepatocytes was investigated. Following the incubation of primary mouse hepatocytes with 100 μM 8-br-cAMP for 8 h, PEPCK mRNA expression was significantly increased, and this increase was significantly suppressed by 2 mM metformin (Fig. 1). This indicates that the experimental conditions are suitable for studying the mechanism of gluconeogenesis. Therefore, the microarray analysis of the lncRNAs expression profile was performed using the same conditions.
Figure 1.
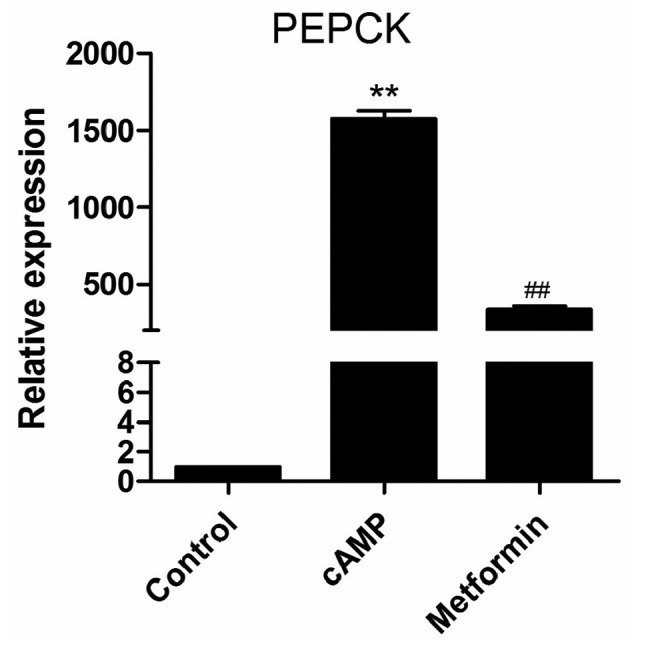
Metformin significantly inhibits cAMP-stimulated PEPCK expression. Primary mouse hepatocytes were incubated with 100 μM 8-br-cAMP in the presence or absence of 2 mM metformin for 8 h. PEPCK mRNA expression was detected by reverse transcription-quantitative polymerase chain reaction. Results are representative of three independent experiments. **P<0.01 vs. control group; ##P<0.01 vs. cAMP group. cAMP, cyclic adenosine monophosphate; PEPCK, phosphoenolpyruvate carboxykinase; 8-br-cAMP, 8-bromoadenosine 3′,5′-cyclic monophosphate.
Profile of the microarray data
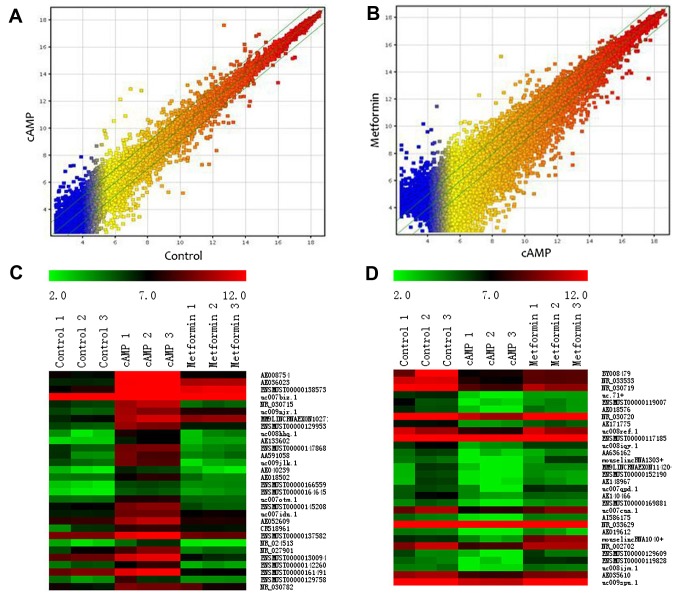
A total of 22,016 lncRNAs were detected in the primary mouse hepatocytes using ArrayStar Mouse LncRNA Microarray version 2.0. The lncRNAs represented in this microarray have been sourced from authoritative databases, including RefSeq, UCSC Knowngenes and Ensembls. Scatter plots were used to assess the variation in the expression of lncRNAs between the cAMP group and the control group (Fig. 2A) or between the metformin group and the cAMP group (Fig. 2B). By setting a threshold for differential expression at fold-change >2.0-fold, 865 differentially expressed lncRNAs (456 upregulated and 409 downregulated lncRNAs) were identified in the cAMP group vs. the control group. Compared with the cAMP group, 4,580 differentially expressed lncRNAs were identified, of which 2,114 lncRNAs were upregulated and 2,466 lncRNAs were downregulated in the metformin group. Among the cAMP-upregulated lncRNAs, 189 were downregulated by metformin. Among the cAMP-downregulated lncRNAs, 167 were upregulated by metformin. Heatmaps demonstrate the lncRNA expression profiles of the groups, and the expression values of the 30 most strongly cAMP-upregulated and cAMP-downregulated lncRNAs (Fig. 2C and D and Table II) reversed by metformin are presented.
Figure 2.
Comparison of lncRNA expression profiles in primary mouse hepatocytes of the three groups. The differential expression profiles of lncRNAs were analyzed by microarray. Scatter plots are presented to show the lncRNA expression variation between (A) the cAMP group and the control group and (B) the metformin group and the cAMP group. The values on the x- and y-axes in the scatter-plot are the normalized signal values of each group (log2 scaled). The lncRNAs above the top green line and below the bottom green line in the scatter plots indicate a >2-fold-change of lncRNA expression. The heatmaps show (C) the top 30 cAMP-upregulated lncRNAs and (D) the top 30 cAMP-downregulated lncRNAs reversed by metformin. Red indicates a relatively high expression and green indicates a relatively low expression. lncRNA, long non-coding RNA; cAMP, cyclic adenosine monophosphate.
Table II.
Top 30 cAMP-upregulated lncRNAs and cAMP-downregulated lncRNAs reversed by metformin.
| Seqname | Fold-change | Regulation (cAMP vs. control) | Fold-change | Regulation (metformin vs. cAMP) |
|---|---|---|---|---|
| AK008754 | 63.58867 | Up | 55.04594 | Down |
| AK036023 | 57.2507 | Up | 3.325115 | Down |
| ENSMUST00000138573 | 37.43524 | Up | 2.130461 | Down |
| uc007biz.1 | 31.35165 | Up | 17.08355 | Down |
| NR_030715 | 25.00611 | Up | 45.54781 | Down |
| uc009njr.1 | 22.53223 | Up | 4.457007 | Down |
| MM9LINCRNAEXON10271− | 19.73751 | Up | 2.071386 | Down |
| ENSMUST00000129953 | 18.7799 | Up | 8.698158 | Down |
| uc008hhq.1 | 15.70811 | Up | 8.776502 | Down |
| AK133602 | 15.02128 | Up | 8.871666 | Down |
| ENSMUST00000147868 | 14.96649 | Up | 32.93101 | Down |
| AA591058 | 14.75416 | Up | 16.43587 | Down |
| uc009jlk.1 | 12.59451 | Up | 43.2916 | Down |
| AK040239 | 8.804657 | Up | 8.449728 | Down |
| AK018502 | 7.718255 | Up | 8.491624 | Down |
| ENSMUST00000166559 | 7.339848 | Up | 2.552899 | Down |
| ENSMUST00000164645 | 6.906824 | Up | 2.483507 | Down |
| uc007otm.1 | 6.888684 | Up | 6.263191 | Down |
| ENSMUST00000145208 | 6.529995 | Up | 19.36338 | Down |
| uc007idu.1 | 6.296531 | Up | 3.856799 | Down |
| AK052609 | 6.237578 | Up | 7.895747 | Down |
| CR518961 | 6.112049 | Up | 2.601747 | Down |
| ENSMUST00000137582 | 6.028889 | Up | 8.330698 | Down |
| NR_024513 | 6.003099 | Up | 10.10774 | Down |
| NR_027901 | 5.708479 | Up | 20.74246 | Down |
| ENSMUST00000130094 | 5.705397 | Up | 33.7096 | Down |
| ENSMUST00000142260 | 5.699138 | Up | 15.27239 | Down |
| ENSMUST00000161491 | 5.59761 | Up | 29.0643 | Down |
| ENSMUST00000129758 | 5.463225 | Up | 5.843725 | Down |
| NR_030782 | 5.312138 | Up | 2.623824 | Down |
| BY008479 | 21.01573 | Down | 2.650546 | Up |
| NR_033533 | 15.91963 | Down | 2.650648 | Up |
| NR_030719 | 14.39216 | Down | 3.259029 | Up |
| uc.71+ | 12.23815 | Down | 2.515803 | Up |
| ENSMUST00000119007 | 8.960969 | Down | 2.669164 | Up |
| AK018576 | 8.671295 | Down | 3.36866 | Up |
| NR_030720 | 6.8741 | Down | 4.131914 | Up |
| AK171775 | 6.527647 | Down | 2.402267 | Up |
| uc008ref.1 | 6.215065 | Down | 5.873268 | Up |
| ENSMUST00000117185 | 6.061 | Down | 3.369349 | Up |
| uc008iqy.1 | 6.043397 | Down | 3.388012 | Up |
| AA636162 | 5.986786 | Down | 2.320948 | Up |
| mouselincRNA1303+ | 5.755683 | Down | 7.49714 | Up |
| MM9LINCRNAEXON11420+ | 5.640478 | Down | 3.268149 | Up |
| ENSMUST00000152190 | 5.534303 | Down | 2.962099 | Up |
| AK148967 | 5.531486 | Down | 3.036806 | Up |
| uc007qpd.1 | 5.508539 | Down | 8.222444 | Up |
| AK140466 | 5.259056 | Down | 2.131494 | Up |
| ENSMUST00000169881 | 5.179452 | Down | 3.134108 | Up |
| uc007cua.1 | 5.058929 | Down | 3.124969 | Up |
| AI586175 | 4.876382 | Down | 4.435117 | Up |
| NR_033629 | 4.820504 | Down | 2.005511 | Up |
| AK019612 | 4.674555 | Down | 2.644345 | Up |
| mouselincRNA1040+ | 4.510026 | Down | 49.62078 | Up |
| NR_002702 | 4.507347 | Down | 4.266528 | Up |
| ENSMUST00000129609 | 4.367185 | Down | 5.31341 | Up |
| ENSMUST00000119828 | 4.262482 | Down | 11.6708 | Up |
| uc008ijm.1 | 4.213697 | Down | 4.486794 | Up |
| AK035610 | 4.009485 | Down | 2.407165 | Up |
| uc009spu.1 | 3.959393 | Down | 2.946273 | Up |
The table lists only some of the results for lncRNAs with an up- or downregulation in expression in the cAMP group compared with the control group and in the metformin group compared with the cAMP group. Seqname, lncRNA name; fold-change, absolute fold-change between the compared groups; up, upregulation; down, downregulation; cAMP, cyclic adenosine monophosphate; lncRNA, long non-coding RNA.
Expression signatures of metformin-reversed lncRNAs
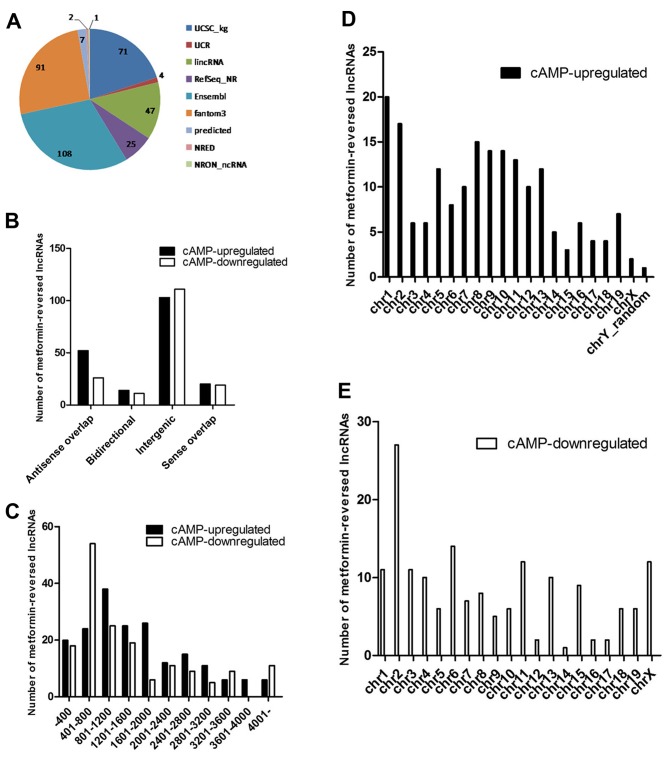
It is likely that these metformin-reversed lncRNAs serve a critical role in the regulation of gluconeogenesis. Therefore, some general signatures of these lncRNAs, such as their sources, classification, length distribution and chromosome distribution, were investigated. A pie chart was constructed to show the number of metformin-reversed lncRNAs collected from different databases (Fig. 3A). There were 78 antisense overlap, 25 bidirectional, 214 intergenic and 39 sense overlap lncRNAs among these lncRNAs (Fig. 3B), which were mainly between 400 and 3,600 nt in length (Fig. 3C). Chromosome distribution analysis demonstrated that these metformin-reversed lncRNAs were located on a number of different chromosomes (Fig. 3D and E).
Figure 3.
Expression signatures of metformin-reversed lncRNAs regulated by cAMP. (A) The numbers of metformin-reversed lncRNAs from various authorative databases. (B) Classification of reversed lncRNAs demonstrated that these lncRNAs were mainly intergenic. (C) The majority of the metformin-reversed lncRNAs were between 400 and 1,200 nucleotides in length. Analysis of chromosome distribution showed the numbers of (D) cAMP-upregulated and metformin-downregulated lncRNAs and (E) cAMP-downregulated and metformin-upregulated lncRNAs located in different chromosomes. lncRNA, long non-coding RNA; cAMP, cyclic adenosine monophosphate.
RT-qPCR validation
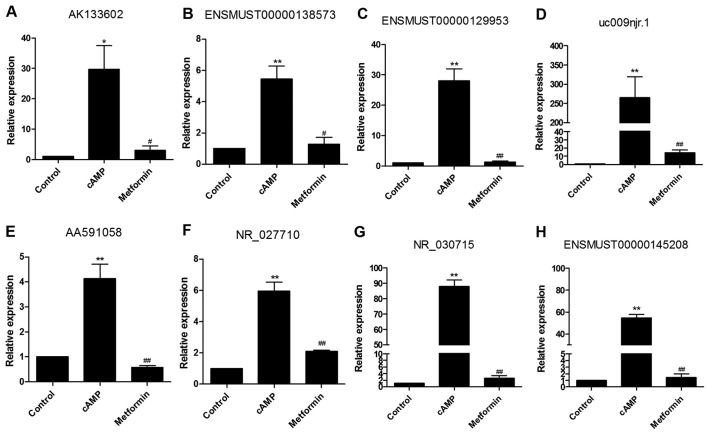
Among the identified metformin-reversed lncRNAs, 8 cAMP-stimulated and metformin-inhibited l ncR NAs (A K1336 02, ENSM UST 0 0 0 0 0138573, ENSMUST00000129953, uc009njr.1, AA591058, NR_027710, NR_030715 and ENSMUST00000145208) were selected for analysis by RT-qPCR in order to verify the microarray data. The expression levels of all 8 cAMP-stimulated lncRNAs were significantly decreased in the presence of metformin as shown by RT-qPCR (Fig. 4), consistent with the results of the microarray analysis.
Figure 4.
Validation results of eight cAMP-upregulated and metformin-downregulated lncRNAs by RT-qPCR are consistent with those of the microarray. Primary mouse hepatocytes were incubated with 100 μM 8-br-cAMP in the presence or absence of 2 mM metformin for 8 h. RT-qPCR demonstrated that cAMP-stimulated (A) AK133602, (B) ENSMUST00000138573, (C) ENSMUST00000129953, (D) uc009njr.1, (E) AA591058, (F) NR_027710, (G) NR_030715 and (H) ENSMUST00000145208 were attenuated by metformin. All results were obtained from three independent experiments. *P<0.05 and **P<0.01 vs. control group; #P<0.05 and ##P<0.01 vs. cAMP group. cAMP, cyclic adenosine monophosphate; lncRNA, long non-coding RNA; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Associated coding gene expression
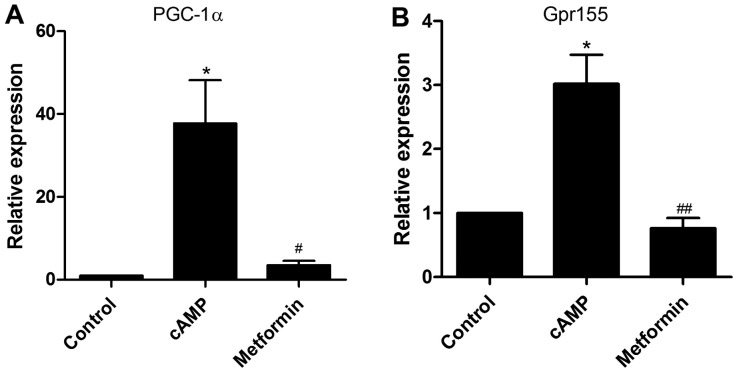
Ponjavic et al (18) emphasized the importance of lncRNA and its adjacent protein-coding gene pairs when investigating the function of lncRNAs. Therefore, using the UCSC genome browser (http://genome.ucsc.edu/) and NONCODE database (http://www.noncode.org), the sequences of the eight validated lncRNAs and their associated coding genes were obtained (data not shown). The lncRNA ENSMUST00000138573 is a 614-nt antisense overlapping lncRNA associated with the G protein-coupled receptor 155 (Gpr155) gene. LncRNA NR_027710 is a sense overlapping lncRNA, which is located near the PGC-1α gene. Notably, RT-qPCR analysis demonstrated that Gpr155 and PGC-1α displayed a similar expression pattern to their associated lncRNAs under the same treatment conditions (Fig. 5A and B). Therefore, it is possible that the two lncRNAs modulate gluconeogenesis through their associated protein-coding genes.
Figure 5.
Expression levels of PGC-1α and Gpr155 are increased by cAMP and are further decreased by metformin treatment. Primary mouse hepatocytes were incubated with 100 μM 8-br-cAMP in the presence or absence of 2 mM metformin for 8 h. (A) PGC-1α and (B) Gpr155 mRNA expression was detected by reverse transcription-quantitative polymerase chain reaction. The experiment was repeated three times with similar results. *P<0.05 vs. control group; #P<0.05 and ##P<0.01 vs. cAMP group. PGC-1α, peroxisome proliferator-activated receptor-γ coactivator-1 1α, Gpr155, G protein-coupled receptor 155; cAMP, cyclic adenosine monophosphate.
Discussion
Excessive glucose output via gluconeogenesis is a critical pathological factor contributing to hyperglycemia in type 2 diabetes mellitus, during which the liver synthesizes glucose from non-carbohydrate precursors, including pyruvate and lactate (19,20). Under the fasting state, increased pancreatic hormone glucagon interacts with the glucagon receptor and activates adenylate cyclase, thus leading to an elevation of intracellular cAMP (10). Elevated cAMP then triggers gluconeogenesis via activation of the transcriptional factor cAMP-response element binding protein (CREB) (10, 21). It is well known that the activity of the gluconeogenic pathway is controlled by the gene expression of several key enzymes, including PEPCK. A large body of evidence from animal studies and diabetic patients has demonstrated that metformin lowers blood glucose levels by inhibiting gluconeogenesis (12–14). The present research team have also observed that metformin decreases glucose production in primary mouse hepatocytes (data not shown). In the present study, cAMP-induced PEPCK mRNA expression was demonstrated to be suppressed by metformin, consistent with previous studies (13,14).
lncRNAs were initially considered to be transcriptional 'noise' of the mammalian genome; however, there is considerable evidence that lncRNAs serve key roles in various biological processes via the regulation of gene expression at the level of chromatin remodeling, transcriptional control and post-transcriptional processing (22). LncRNAs have also been implicated in numerous human diseases, including Alzheimer's disease (23), cardiovascular diseases (24), diabetes (25) and various cancers (26). There are many studies demonstrating that lncRNAs are involved in the differentiation and homeostasis of metabolic tissues, including islets, skeletal muscle and adipose tissues, as summarized in a recent review (27). Particularly in islets, there is growing evidence that lncRNAs are involved in the determination of β-cell identity and participate in the misregulation of gene expression during type 2 diabetes (7,28). However, to the best of our knowledge, the effect of lncRNA on hepatic gluconeogenesis has not been elucidated. In the present study, a hepatocyte model with cAMP and metformin treatment was used to better understand the potential role of lncRNAs in the hepatic gluconeogenesis process.
Using microarray analysis, 865 and 4,580 differentially expressed lncRNAs were identified in the cAMP group vs. the control group and the metformin group vs. the cAMP group, respectively. Among the cAMP-regulated lncRNAs, 356 were reversed by metformin treatment, and the expression of 189 AMP-upregulated lncRNAs was decreased by metformin. Previous studies have demonstrated that metformin suppresses the cAMP-stimulated expression of genes involved in gluconeogenesis (14,29). By analyzing the expression signatures of the metformin-reversed lncRNAs, it was found that the majority were long intergenic noncoding RNAs. Therefore, it is possible that these lncRNAs regulate the expression of neighbouring protein-coding genes. A set of 8 cAMP-upregulated and metformin-downregulated lncRNAs was selected with which to validate the results of the micro-array using RT-qPCR. The results suggest that metformin may decrease gluconeogenesis by globally altering hepatic lncRNA expression.
Although the precise roles of these lncRNAs in gluconeogenesis remain unknown, lncRNAs have been demonstrated to regulate neighbouring gene expression through cis- and trans-mechanisms (5). Notably, a strong co-expression of lncRNA NR_027710 and PGC-1α was validated by the RT-qPCR analysis in the present study, as shown in Figs. 4F and 5A. This implies a potential role of NR_027710 in the regulation of PGC-1α expression. Hormonal signaling regulates hepatic gluconeogenesis by triggering a cascade of transcriptional events involving various transcriptional activators and coactivators, including PGC-1α, CREB and transducer of regulated CREB activity 2 (30–32). LncRNA NR_027710 has been found to be transcribed from the natural-sense strand of the gene PGC-1α, which encodes the transcriptional coactivator PGC-1α. Herzig et al (30) demonstrated that PGC-1α is induced by CREB to trigger the expression of gluconeogenic genes. The results of the present study and previous studies (14,17) demonstrated that 8-br-cAMP caused a significant increase in the PGC-1α transcript, which was suppressed by metformin. According to the bioinformatic analysis, ENSMUST00000138573, a 614-nt lncRNA exhibits a natural antisense association with the coding gene Gpr155 in chromosome 2. In the present study, the expression level of Gpr155 displayed a similar pattern of change to that of lncRNA ENSMUST00000138573. Kobayashi et al (33) revealed that Gpr155 was one of four candidate genes for type 2 diabetes by performing exome sequencing analysis. It has been shown that lncRNAs act as transcriptional cofactors to modulate the transcription of adjacent protein coding genes (34), which suggests that lncRNA NR_027710 and lncRNA ENSMUST00000138573 may affect the expression of PGC-1α and Gpr155 through an unknown mechanism. These two lncRNA-mRNA gene pairs highlight the importance of lncRNAs in the modulation of glucose homeostasis.
In conclusion, to the best of our knowledge, the present study is the first to reveal the role of lncRNA in gluconeogenesis by conducting a global microarray analysis. It is likely that metformin inhibits hepatic gluconeogenesis by regulating lncRNA expression. In addition, two lncRNAs NR_027710 and ENSMUST00000138573 are associated with PGC-1α and Gpr155, respectively. The two pairs of lncRNAs and coding protein genes exhibit a similar expression pattern, suggesting that lncRNAs may exert their functions through interactions with coding transcripts and proteins in gluconeogenesis. The present study provides a new perspective for understanding the glucose-lowering mechanism of metformin. However, the exact regulatory mechanisms of the specific lncRNAs involved in gluconeogenesis require further investigation, potentially via loss and gain of function studies.
Acknowledgments
The present study was funded by grants from the National Natural Science Foundation of China (grant nos. 81170720, 81270910, 81370876 and 81471030).
Glossary
Abbreviations
- lncRNA
long non-coding RNA
- cAMP
cyclic adenosine monophosphate
- PGC-1α
peroxisome proliferator-activated receptor-γ coactivator-1α
- Gpr155
G protein-coupled receptor 155
- 8-br-cAMP
8-bromoadenosine 3′,5′-cyclic monophosphate
- RT-qPCR
reverse transcription-quantitative polymerase chain reaction
- PEPCK
phosphoenolpyruvate carboxykinase
- CREB
cAMP-response element binding protein
References
- 1.Ning G. Decade in review-type 2 diabetes mellitus: At the centre of things. Nat Rev Endocrinol. 2015;11:636–638. doi: 10.1038/nrendo.2015.147. [DOI] [PubMed] [Google Scholar]
- 2.Kitamura Y, Accili D. New insights into the integrated physiology of insulin action. Rev Endocr Metab Disord. 2004;5:143–149. doi: 10.1023/B:REMD.0000021436.91347.93. [DOI] [PubMed] [Google Scholar]
- 3.Accili D. Lilly lecture 2003: The struggle for mastery in insulin action: From triumvirate to republic. Diabetes. 2004;53:1633–1642. doi: 10.2337/diabetes.53.7.1633. [DOI] [PubMed] [Google Scholar]
- 4.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: Insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 5.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morán I, Akerman I, van de Bunt M, Xie R, Benazra M, Nammo T, Arnes L, Nakić N, García-Hurtado J, Rodríguez-Seguí S, et al. Human β cell transcriptome analysis uncovers lncRNAs that are tissue-specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell Metab. 2012;16:435–448. doi: 10.1016/j.cmet.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun L, Goff LA, Trapnell C, Alexander R, Lo KA, Hacisuleyman E, Sauvageau M, Tazon-Vega B, Kelley DR, Hendrickson DG, et al. Long noncoding RNAs regulate adipogenesis. Proc Natl Acad Sci USA. 2013;110:3387–3392. doi: 10.1073/pnas.1222643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao Y, Wu F, Zhou J, Yan L, Jurczak MJ, Lee HY, Yang L, Mueller M, Zhou XB, Dandolo L, et al. The H19/let-7 double-negative feedback loop contributes to glucose metabolism in muscle cells. Nucleic Acids Res. 2014;42:13799–13811. doi: 10.1093/nar/gku1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pilkis SJ, el-Maghrabi MR, Claus TH. Hormonal regulation of hepatic gluconeogenesis and glycolysis. Annu Rev Biochem. 1988;57:755–783. doi: 10.1146/annurev.bi.57.070188.003543. [DOI] [PubMed] [Google Scholar]
- 11.Chan JC, Deerochanawong C, Shera AS, Yoon KH, Adam JM, Ta VB, Chan SP, Fernando RE, Horn LC, Nguyen TK, et al. Role of metformin in the initiation of pharmacotherapy for type 2 diabetes: An Asian-Pacific perspective. Diabetes Res Clin Pract. 2007;75:255–266. doi: 10.1016/j.diabres.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 12.Inzucchi SE, Maggs DG, Spollett GR, Page SL, Rife FS, Walton V, Shulman GI. Efficacy and metabolic effects of metformin and troglitazone in type II diabetes mellitus. N Engl J Med. 1998;338:867–872. doi: 10.1056/NEJM199803263381303. [DOI] [PubMed] [Google Scholar]
- 13.Shaw RJ, Lamia KA, Vasquez D, Koo SH, Bardeesy N, Depinho RA, Montminy M, Cantley LC. The kinase LKB1 mediates glucose homeostasis in liver and therapeutic effects of metformin. Science. 2005;310:1642–1646. doi: 10.1126/science.1120781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foretz M, Hébrard S, Leclerc J, Zarrinpashneh E, Soty M, Mithieux G, Sakamoto K, Andreelli F, Viollet B. Metformin inhibits hepatic gluconeogenesis in mice independently of the LKB1/AMPK pathway via a decrease in hepatic energy state. J Clin Invest. 2010;120:2355–2369. doi: 10.1172/JCI40671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Madiraju AK, Erion DM, Rahimi Y, Zhang XM, Braddock DT, Albright RA, Prigaro BJ, Wood JL, Bhanot S, MacDonald MJ, et al. Metformin suppresses gluconeogenesis by inhibiting mitochondrial glycerophosphate dehydrogenase. Nature. 2014;510:542–546. doi: 10.1038/nature13270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Yoon JC, Puigserver P, Chen G, Donovan J, Wu Z, Rhee J, Adelmant G, Stafford J, Kahn CR, Granner DK, et al. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature. 2001;413:131–138. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]
- 18.Ponjavic J, Oliver PL, Lunter G, Ponting CP. Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet. 2009;5:e1000617. doi: 10.1371/journal.pgen.1000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nordlie RC, Foster JD, Lange AJ. Regulation of glucose production by the liver. Annu Rev Nutr. 1999;19:379–406. doi: 10.1146/annurev.nutr.19.1.379. [DOI] [PubMed] [Google Scholar]
- 20.Pilkis SJ, Granner DK. Molecular physiology of the regulation of hepatic gluconeogenesis and glycolysis. Annu Rev Physiol. 1992;54:885–909. doi: 10.1146/annurev.ph.54.030192.004321. [DOI] [PubMed] [Google Scholar]
- 21.Gonzalez GA, Montminy MR. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell. 1989;59:675–680. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- 22.Lee JT, Bartolomei MS. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. 2013;152:1308–1323. doi: 10.1016/j.cell.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 23.Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G, III, Kenny PJ, Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Archer K, Broskova Z, Bayoumi AS, Teoh JP, Davila A, Tang Y, Su H, Kim IM. Long non-coding RNAs as master regulators in cardiovascular diseases. Int J Mol Sci. 2015;16:23651–23667. doi: 10.3390/ijms161023651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ding GL, Wang FF, Shu J, Tian S, Jiang Y, Zhang D, Wang N, Luo Q, Zhang Y, Jin F, et al. Transgenerational glucose intolerance with Igf2/H19 epigenetic alterations in mouse islet induced by intrauterine hyperglycemia. Diabetes. 2012;61:1133–1142. doi: 10.2337/db11-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang G, Lu X, Yuan L. LncRNA: A link between RNA and cancer. Biochim Biophys Acta. 2014;1839:1097–1109. doi: 10.1016/j.bbagrm.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 27.Zhao XY, Lin JD. Long noncoding RNAs: A new regulatory code in metabolic control. Trends Biochem Sci. 2015;40:586–596. doi: 10.1016/j.tibs.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fadista J, Vikman P, Laakso EO, Mollet IG, Esguerra JL, Taneera J, Storm P, Osmark P, Ladenvall C, Prasad RB, et al. Global genomic and transcriptomic analysis of human pancreatic islets reveals novel genes influencing glucose metabolism. Proc Natl Acad Sci USA. 2014;111:13924–13929. doi: 10.1073/pnas.1402665111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao J, Meng S, Chang E, Beckwith-Fickas K, Xiong L, Cole RN, Radovick S, Wondisford FE, He L. Low concentrations of metformin suppress glucose production in hepatocytes through AMP-activated protein kinase (AMPK) J Biol Chem. 2014;289:20435–20446. doi: 10.1074/jbc.M114.567271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herzig S, Long F, Jhala US, Hedrick S, Quinn R, Bauer A, Rudolph D, Schutz G, Yoon C, Puigserver P, et al. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature. 2001;413:179–183. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- 31.Puigserver P, Rhee J, Donovan J, Walkey CJ, Yoon JC, Oriente F, Kitamura Y, Altomonte J, Dong H, Accili D, et al. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature. 2003;423:550–555. doi: 10.1038/nature01667. [DOI] [PubMed] [Google Scholar]
- 32.Koo SH, Flechner L, Qi L, Zhang X, Screaton RA, Jeffries S, Hedrick S, Xu W, Boussouar F, Brindle P, et al. The CREB coactivator TORC2 is a key regulator of fasting glucose metabolism. Nature. 2005;437:1109–1111. doi: 10.1038/nature03967. [DOI] [PubMed] [Google Scholar]
- 33.Kobayashi M, Ohno T, Ihara K, Murai A, Kumazawa M, Hoshino H, Iwanaga K, Iwai H, Hamana Y, Ito M, et al. Searching for genomic region of high-fat diet-induced type 2 diabetes in mouse chromosome 2 by analysis of congenic strains. PLoS One. 2014;9:e96271. doi: 10.1371/journal.pone.0096271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20:1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]