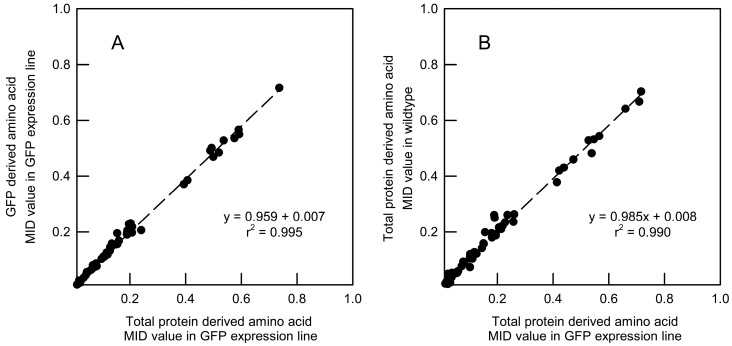
Figure 5.
Validation of GFP as a reporter protein for csMFA. (A) Comparison of MIDs of amino acids derived from either GFP or total protein obtained from the roots of A. thaliana seedlings expressing GFP. The seedlings were grown in 20% [13C6]glucose and total ion counts measured for amino acid mass peaks were processed into MIDs to obtain average data on each mass peak for each fragment. The plot includes data from 12 amino acids, corresponding to the reliable detection of identical fragments in the two protein fractions. A paired t-test established that there was no significant difference between the abundances of individual amino acid isotopomers derived from total protein and GFP (t-value = 0.063, d.f. = 68, p = 0.950). (B) Comparison of total protein MIDs from wild-type and GFP-expressing seedlings. In both plots, the coefficient of linear regression was one with an intercept of zero and the coefficient of determination was close to unity demonstrating the equivalence of the mass isotopomer distributions in the systems under comparison. A paired t-test established that there was no significant difference between the abundances of individual amino acid isotopomers derived from total protein in wild-type and the GFP-expressing line (t-value = 0.060, d.f. = 59, p = 0.953).