Fig. 3. Method for detection of two mutations separated in time.
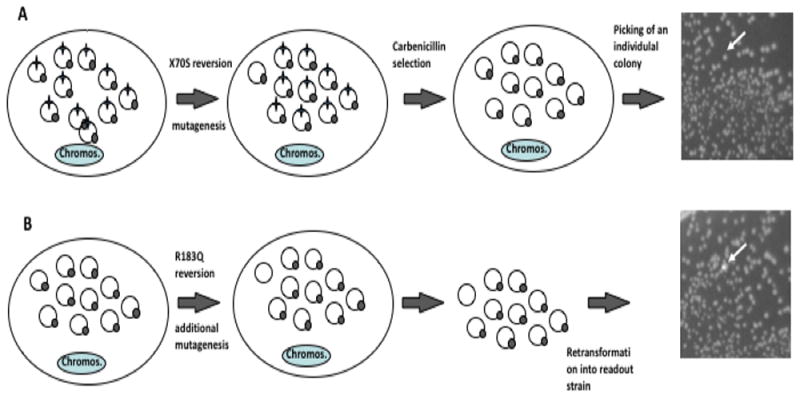
Colonies representing reversions in TEM1 β-lactamase are expanded under restrictive (mutagenic) conditions, their plasmid pools recovered through miniprep and retransformed. The two reversion reporters are shown as star (S70X) and circle (Q183R). A. Detection of first mutation: once reversion at the S70X site occurs, under selective pressure, the reversion events get amplified, representing a majority of the plasmid population, and leading to carbenicillin resistance. B. Detection of second mutation: single, carbenicillin-resistant colonies (white arrow) are grown. Plasmids from these cultures are recovered. Reversions at the Q183 site of the GFP reporter are detected by retransformation of recovered plasmids into a readout strain, producing fluorescent colonies on a background on non-fluorescent ones (white arrow). Reversion cannot have already been present in one of the copies of the plasmid pool when the first mutation occurred if the frequency of reversion is lower than one divided by the copy number of the reporter plasmid (in this example, one in 10).
