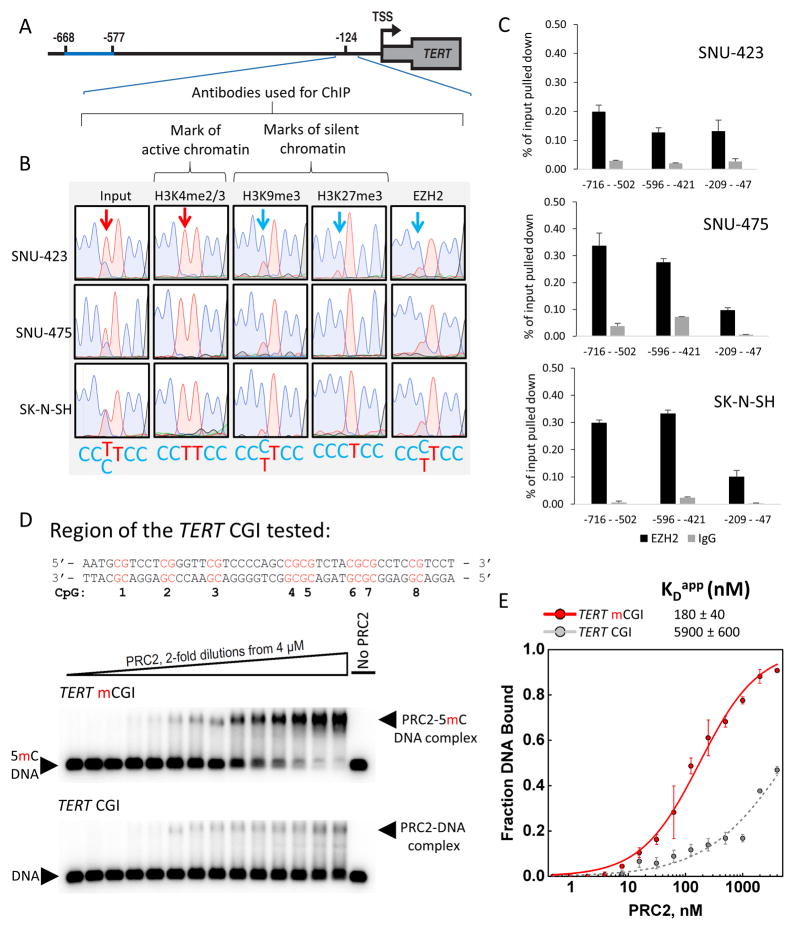
Figure 3. PRC2 Displays a Strong Binding Preference for 5mC-Rich TERT Alleles in vivo in −124 Cells and Methylated TERT CGI DNA in vitro.
(A) Schematic illustrating the position of the allele-specific ChIP analysis (−209 – −47).
(B) Representative Sanger sequencing traces from DNA isolated by ChIP and amplified by three PCR reactions pooled prior to sequencing, showing the presence of H3K27me3, H3K9me3 and EZH2 on the inactive allele in HCC (SNU-423, SNU-475) and neuroblastoma (SK-N-SH) −124 heterozygous tumor-derived cell lines. Arrows indicate the position of the heterozygous −124 mutation.
(C) Quantitative assessment of EZH2 occupancy of TERT promoters in three −124 mutant cell lines. Data are + SEM, n=3 technical replicates.
(D) Sequence tested for PRC2 binding in vitro. CpG numbers correspond to those listed in Fig. 2D. Representative EMSA gels showing binding of purified recombinant human PRC2 to the fully CpG-methylated TERT DNA vs. the same sequence lacking 5mC modifications.
(E) Binding data fit with equilibrium binding curves, error bars are ± SD (n=3 independent experiments).