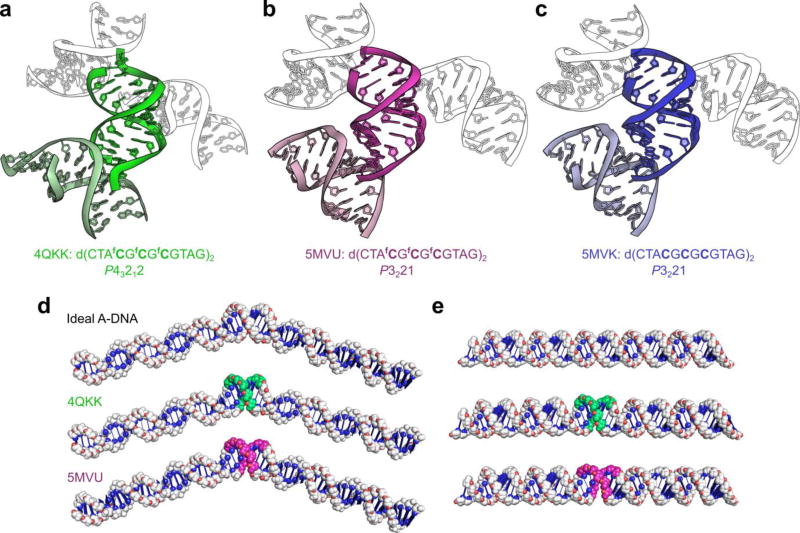
Figure 2. Crystal packing interactions exert a greater effect on DNA conformation than 5-formylcytosine in the crystalline state.
(a–c) Cartoons illustrating the crystal packing of an identical 5-formylcytosine-containing sequence crystallized in different space groups (4QKK & 5MVU), and that of its unmodified analogue 5MVK. (a) The duplex d(CTAfCGfCGfCGTAG)2, when crystallized in the P43212 space group from a buffer containing 1.8 M lithium sulfate,17 interacts strongly with neighboring duplexes within the crystal, resulting in pronounced kinking of the backbone at the duplex ends and consequently deviation from A-type geometry at the terminal nucleotides; however, the duplex’s core, which contains all of the fCpG repeats, adopts the A-form conformation. (b) The same fC-containing duplex, when crystallized in the P3221 space group under milder conditions (<100 mM salt), does not exhibit significant deviation from A-type geometry. (c) The duplex d(CTACGCGCGTAG)2, which also crystallizes in the P3221 space group under milder conditions, is very similar in conformation to its fC-containing counterpart when crystallized isomorphously (rmsd of 0.65 Å, 486 atoms) and forms similar crystal contacts. Note that some symmetry mates have been omitted for clarity. (d) Modelling of B-DNA 60-bp duplexes containing a central 8-bp region of either ideal A-DNA or the region d(AfCGfCGfCGT)2 from the crystal structures 4QKK and 5MVU (fC∙G base pairs shown in green and magenta, respectively). Alterations in the helical trajectory of B-DNA caused by the fC-containing structures closely resemble those of an ideal A-B-DNA junction. (e) Modelling of A-DNA 60-mers containing the 8-bp regions of the aforementioned crystal structures. The fC-containing structures, when flanked by ideal A-DNA, are almost indistinguishable from a continuous stretch of ideal A-DNA. See Supplementary Fig. 4 for an extended comparison.