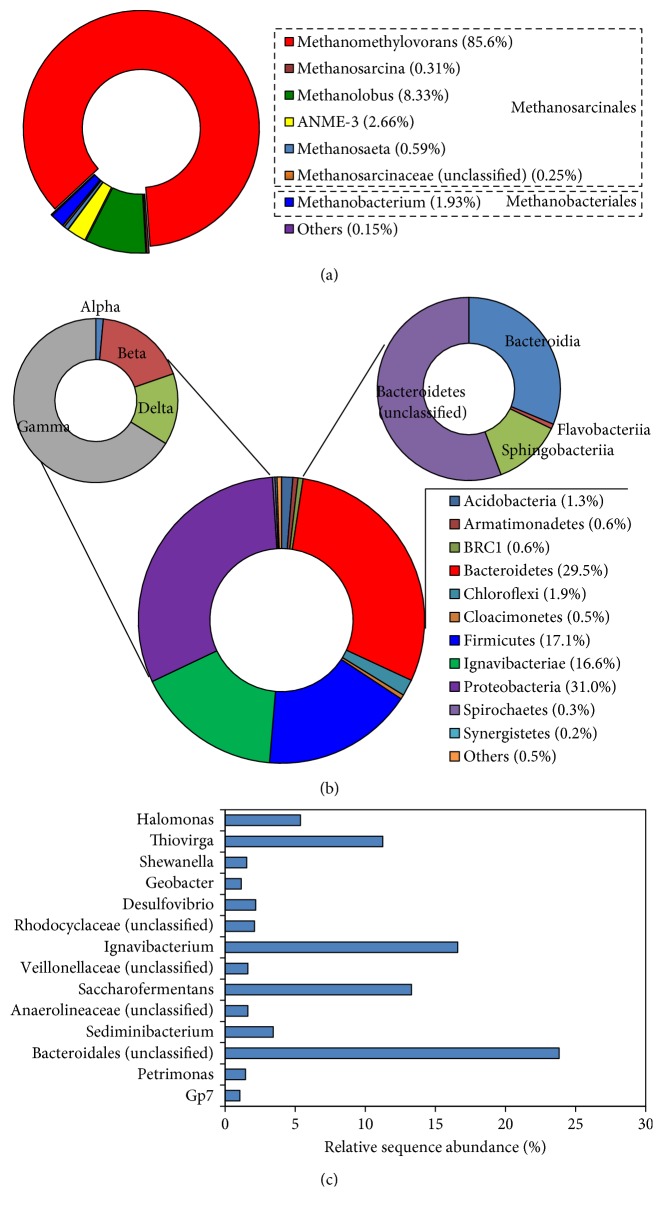
Figure 1.
High-throughput sequencing analysis of archaeal and bacterial 16S rRNA sequences of sludge samples taken from a tetramethylammonium-degrading CMSS reactor. (a) Phylogenetic distribution of the detected archaeal populations. “Others” included the low abundant populations, namely, Methanolinea (0.01%), Methanospirillum (0.01%), Methanobacteriaceae (unclassified) (0.04%), and Thermoplasmata (0.09%). (b) Phylogenetic distribution of the detected bacterial populations at the phylum and class levels. “Others” included 10 bacterial phylum taxa with each abundance <0.05%. (c) Distribution of bacterial genera with relative sequence abundance >1%.