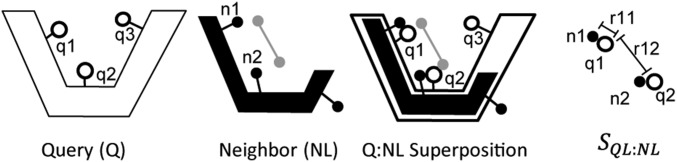
Fig. 4.
Calculating LBias SIM score. A query protein Q is shown at Left with three atoms, q1, q2, and q3, identified specifically. The second panel shows a ligand-containing protein NL, structurally similar to Q with the ligand shown as a gray line connecting two gray atoms. Ligand-binding residues, n1 and n2, are identified. The NL complex is superposed onto Q. Residues from Q that interact with the superposed ligand are identified (q1 and q2 in the above). A protein–ligand similarity score between QL and NL (SQL:NL) is then calculated, where . SQL:NL is a function of all of the pairwise distances (e.g., r11, r12) between the atoms from Q interacting with the superposed ligand and the atoms from N interacting with the native ligand if the two atoms in question make chemically similar contacts with the ligands. For example, if n1 makes a hydrogen bond with the ligand, but q1 is hydrophobic, m11 would be zero and there would be no contribution to SQL:NL from this pair of atoms (Materials and Methods).