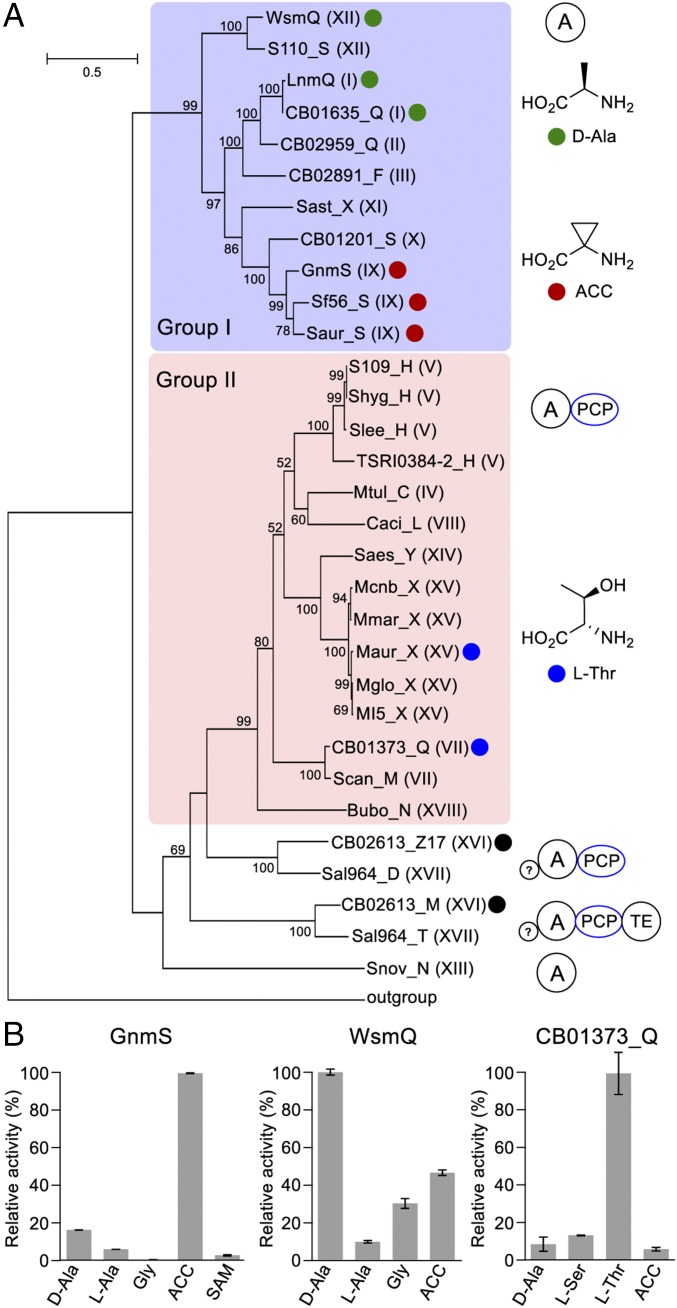
Fig. 3.
Functional diversity of the A proteins or domains from LNM-type biosynthetic machineries. (A) Phylogenetic analysis of the A proteins from the 28 LNM-type machineries, in comparison with LnmQ, which specifies d-Ala (29), revealing two major groups. AfsK (BAA08229) from Streptomyces coelicolor was used as the outgroup. Roman numerals in parentheses refer to the corresponding clades shown in Fig. 2A. The architectures of A proteins from different groups are shown (Right), with A as a discrete protein (group I), A-PCP didomain (group II), and A-PCP accompanied by an extra N-terminal sequence (?) with/without an additional C-terminal thioesterase domain (the rest). The colored dots denote A proteins whose substrate specificities have been confirmed experimentally or deduced from the isolated natural products: green, d-Ala; red, ACC; blue, l-Thr; black, preferred substrate not detected among the 22 amino acids tested (also see SI Appendix, Table S37 for substrate specificities predicted based on the NRPS codes). (B) In vitro assay of representative A proteins to determine their substrate specificities, as exemplified by GnmS, WsmQ, and CB01373_Q that specify ACC, d-Ala, and l-Thr, respectively (also see SI Appendix, Fig. S29). Error bars are generated from three replicates.