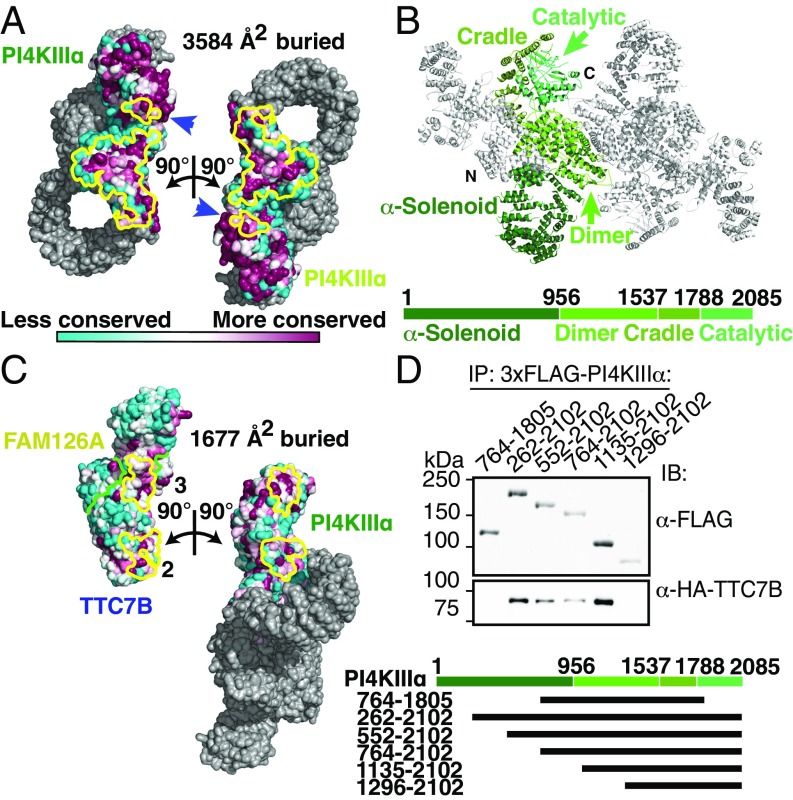
Fig. 2.
PI4KIIIα homodimerizes and interacts stably with TTC7 via large conserved surfaces. (A) Solvent-accessible surface representation of PI4KIIIα/TTC7B/FAM126A complex, with the PI4KIIIα surface (excluding the α-solenoid) colored by conservation. Each heterotrimer is rotated by ±90° as indicated to reveal the homodimerization interface. PI4KIIIα surface residues participating in the dimerization are outlined in yellow. Blue arrows indicate regions of the kinase catalytic domains that contact the opposite copy of PI4KIIIα. (B) Ribbon representation of the PI4KIIIα complex, with one copy of PI4KIIIα colored by domain. Boundary residue numbers for each domain are indicated on the schematic below. (C) Solvent-accessible surface representation of PI4KIIIα/TTC7B/FAM126A complex, with the TTC7/FAM126 complex and PI4KIIIα surfaces colored by conservation, as in A. PI4KIIIα and the TTC7B/FAM126A complex are each rotated by ±90° as indicated to reveal the PI4KIIIα/TTC7B dimerization interface. Interacting residues are outlined in yellow, while the TTC7B/FAM126A boundary is indicated by a green line. TTC7 surfaces 2 and 3 interact with the dimerization and cradle domains of PI4KIIIα, respectively. The additional contact between TTC7B and the tip of the PI4KIIIα α-solenoid, which was not sufficiently resolved to model by a Cα trace, is neither shown nor included in the surface area calculation. (D) Truncation constructs of 3xFLAG-tagged PI4KIIIα were overexpressed alongside HA-TTC7B and EGFP-FAM126A(2–289) in Expi293 cells. α-FLAG-immunoprecipitated samples were resolved by SDS/PAGE and immunoblotted with anti-FLAG and anti-HA antibodies to probe coprecipitation of TTC7B with the different PI4KIIIα constructs. Boundaries of PI4KIIIα truncation constructs are indicated above the blot and in the schematic at bottom.