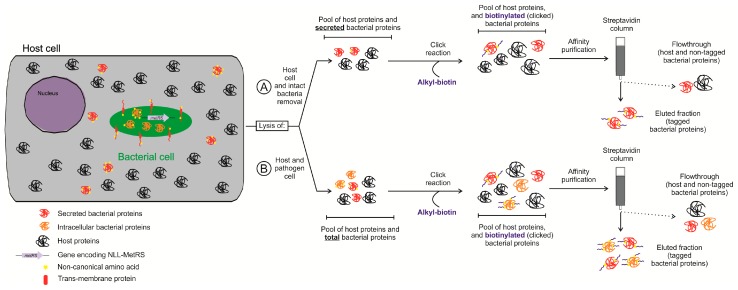
Figure 3.
Bio-orthogonal labeling and purification of labeled bacterial (secreted) proteins from infected host cells. The scheme depicts the workflow for bio-orthogonal tagging of bacterial proteins with non-natural amino acids followed by click chemistry, and isolation of labeled pathogen proteins. Host cells are infected with bacteria, non-natural amino acids are added to the medium prior to infection (pre-labeled bacteria), or upon infection. Bacteria expressing the mutant methionyl-tRNA synthetase (NLL-MetRS), can incorporate non-natural Met analogous exclusively into bacterial proteins. Newly synthetized bacterial proteins will thus incorporate this non-natural Met. After infection, host cells are lysed either in (A) a selective manner keeping bacterial cells intact (e.g., to study the bacterial secretome) or (B) by complete lysis enabling to study the proteome of the pathogen and host simultaneously during infection in a more global manner. Particularly, in the case of (A), bacterial cells are typically removed by centrifugation before downstream processing of the obtained lysate. Subsequently, proteins are clicked using alkyl-biotin (blue). In this step, only proteins that have incorporated non-natural amino acid will be conjugated to biotin. Finally, a purification step using streptavidin affinity chromatography is performed. Here, non-tagged proteins do not bind the streptavidin resin and are thus easily removed, while the biotinylated (clicked) proteins can be recovered. Thus, bacterial (secreted) proteins are highly enriched in the sample and can be studied to understand changes in the secretome (A) or in the bacterial proteome (B) of a bacterial pathogen during infection.