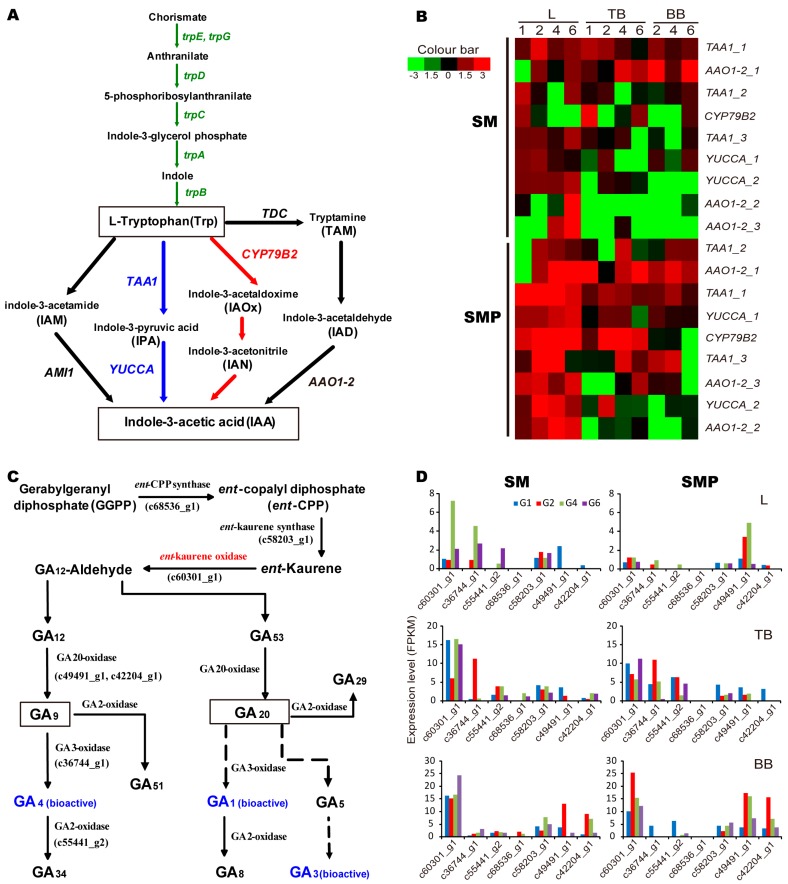
Figure 5.
Genes involved in IAA and GA biosynthesis pathways in willow. (a) Pathway of IAA biosynthesis. Green arrows, tryptophan synthetic pathway; blue and red arrows, steps for which gene and enzymatic function are known in the tryptophan-dependent IAA biosynthetic pathway; letters in italics, genes involved in the conversion process. (b) Heat map of genes involved in IAA biosynthesis during development. Scaled log2 expression values are shown from green to red, indicating low to high expression. TAA1, L-tryptophan-pyruvate aminotransferase; AAO1_2, indole-3-acetaldehyde oxidase; YUCCA, indole-3-pyruvate monooxygenase; CYP79B2, tryptophan N-monooxygenase. L, leaf; TB, top branch; BB, basal branch; SM, S. matsudana; SMP, S. matsudana var. pseudo-matsudana. (c) Gibberellin biosynthesis pathways. (d) Expression patterns of genes involved in gibberellin biosynthesis during development.