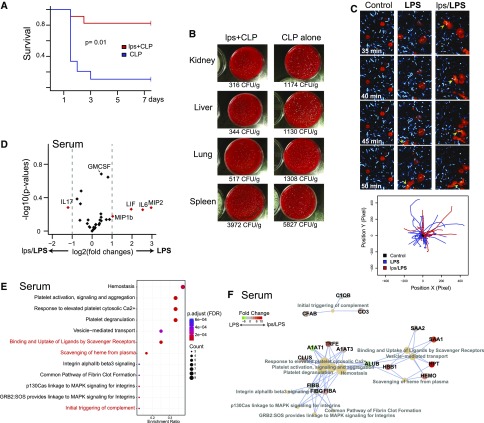
Figure 1.
Endotoxin preconditioning improves survival, reduces inflammation, and maintains scavenging ability. (A) Male C57BL/6 mice were subjected to CLP alone (blue; nonpreconditioned), or to 0.25 mg/kg endotoxin (E. coli serotype 0111:B4) followed 24 hours later by CLP (red; preconditioned), and survival was determined at indicated time points (n=8 per group). The preconditioning dose of endotoxin is denoted as lps in order to distinguish it from high-dose endotoxin (LPS, 5 mg/kg), which is used in subsequent experiments. The CLP alone group received an equivalent dose of sterile normal saline vehicle. P=0.01 by Cox proportional hazards model likelihood ratio. (B) Indicated organs were harvested 24 hours after CLP and bacterial count, estimated from CFU was adjusted for each organ’s weight (n=2 per group with triplicates for each dish). (C) Peritoneal macrophages (red; CellTracker) were incubated with live E. coli (cyan; CFP), and time-lapse images were obtained to assess phagocytic activities. These macrophages were harvested from C57BL/6 mice 12 hours after saline vehicle (control), LPS 5 mg/kg (LPS), or lps 0.25 mg/kg followed 24 hours later by 5 mg/kg administered intraperitoneally (lps/LPS). Arrowheads follow specific mobile macrophages with phagocytized E. coli (yellow) over time. Tracking of cells was performed using a custom plugin for ImageJ.18 Although macrophages from unstimulated mice showed minimal activity, macrophages from LPS and lps/LPS groups exhibited high mobility and phagocytosis. Original magnification ×60. (D) Volcano plot of serum cytokine/chemokine multiplex panel. The serum cytokine/chemokine levels were measured 4 hours after 5 mg/kg LPS administered intraperitoneally. Positive-value log2 fold changes on the x-axis indicate LPS (nonpreconditioned) > lps/LPS (preconditioned) (n=3 per group). (E and F) Differentially expressed serum proteins and their associated pathways are shown (LPS alone versus lps/LPS; Reactome pathway analysis37). Proteins were identified using two-dimensional difference gel electrophoresis in tandem with mass spectrometry (Supplemental Figure 1A). Protein extracts from serum were collected 4 hours after the 5 mg/kg LPS dose for both nonpreconditioned and preconditioned animals. Three samples were pooled for each experimental condition.