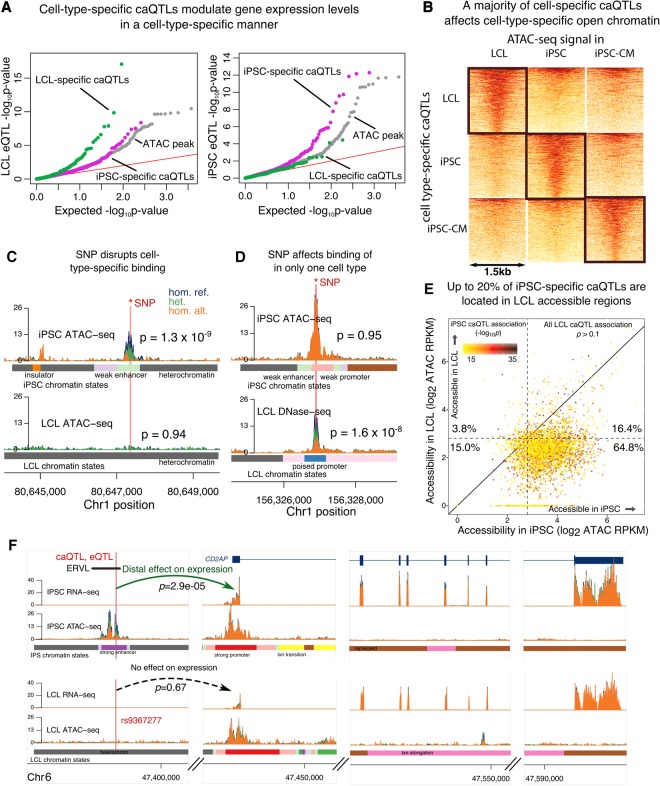
Figure 2.
Mechanisms of cell-type–specific regulatory variation. (A) QQ-plot of LCL and iPSC eQTL signal conditioned on LCL- and iPSC-specific caQTLs. Higher enrichment of LCL (iPSC) eQTLs among LCL (iPSC) caQTLs links cell-type–specific regulation of chromatin accessibility to cell-type–specific regulation of gene expression. (B) Chromatin accessibility signal around cell-specific caQTLs in corresponding cell types (black rectangles) and in other cell types. A lack of accessibility in other cell types suggests that cell-specific caQTLs often affect cell-specific accessible regions, e.g., C. (C,D) Examples of cell-type–specific regulatory effects of genetic variation. SNP is correlated with accessibility of an iPSC-specific open chromatin region in iPSCs only (C) or of a nonspecific open chromatin region in LCLs only (D). (E) Scatter plot of iPSC and LCL chromatin accessibility at iPSC-specific caQTLs. About 20% of iPSC-specific caQTLs are accessible in LCLs. Plot of LCL-specific caQTLs in Supplemental Figure S15. (F) Example of an iPSC-specific caQTL that is also an iPSC-specific eQTL. SNP rs9367277 is associated with both chromatin accessibility of a strong enhancer and with expression of the CD2AP gene in iPSCs. Interestingly, rs9367277 lies in a transposable element of the ERVL family, which is preferentially activated in embryonic stem cells (Kunarso et al. 2010).