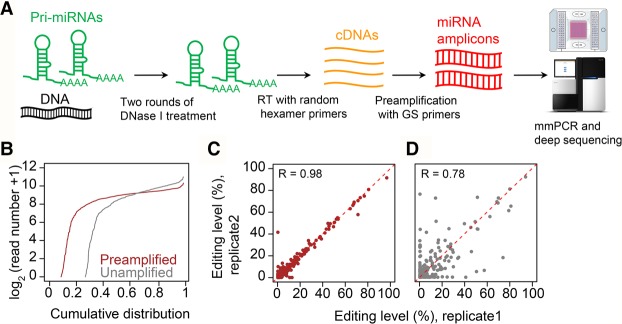
Figure 1.
The development and performance of miR-mmPCR-seq. (A) Schematic diagram of miR-mmPCR-seq. First, multiplex primers covering the pre-miRNA hairpin and flanking 70–150 bp regions were designed. Forty-eight preamplified cDNA samples and 48 pools of primers were loaded to the Fluidigm microfluidic chip. The PCR products from the same cDNA sample were automatically pooled. For each sample, the PCR products were barcoded and pooled together, and subject to 150-bp paired-end sequencing: (RT) reverse transcription; (GS) gene-specific. (B) Comparison of the cumulative distribution of amplicon coverage between unamplified and preamplified cDNA samples. The y-axis shows the number of reads in log2 scale. Read numbers are normalized to 0.25 million mapped reads per sample. (C,D) The reproducibility of RNA editing levels measured using preamplified and unamplified cDNA samples for sites with at least 50 reads in both samples. The color codes are the same as in B.