Fig. 3. RT-PCR analysis of correction of reading frame.
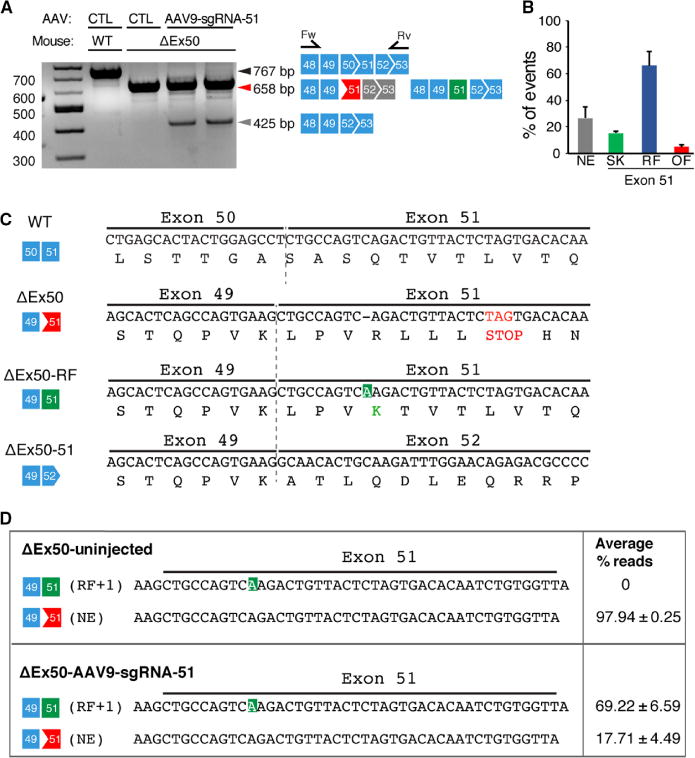
(A) RT-PCR of RNA from the tibialis anterior muscles of WT and ΔEx50 mice 3 weeks after intramuscular injection of the AAV9-sgRNA-51 and AAV9-Cas9 expression vectors. Lower dystrophin bands indicate deletion of exon 51. Primer positions in exons 48 and 53 are indicated. (B) Percentage of events detected at exon 51 after AAV9-Cas9/sgRNA-51 treatment using RT-PCR sequence analysis of TOPO-TA (topoisomerase-based thymidine to adenosine) generated clones. For each of four different samples, we generated and sequenced 40 clones. RT-PCR products were divided into four groups: Not edited (NE), exon 51–skipped (SK), reframed (RF), and out of frame (OF). (C) Sequence of the RT-PCR products of the ΔEx50–51 mouse dystrophin lower band confirmed that exon 49 spliced directly to exon 52, excluding exon 51. Sequence of RT-PCR products of ΔEx50 reframed (ΔEx50-RF) is also shown. (D) Deep-sequencing analysis of RT-PCR products from the upper band containing ΔEx50-NE and ΔEx50-RF was shown. Sequence of RT-PCR products revealing insertions (highlighted in green) is also depicted. n = 4. Data are represented as means ± SEM.
