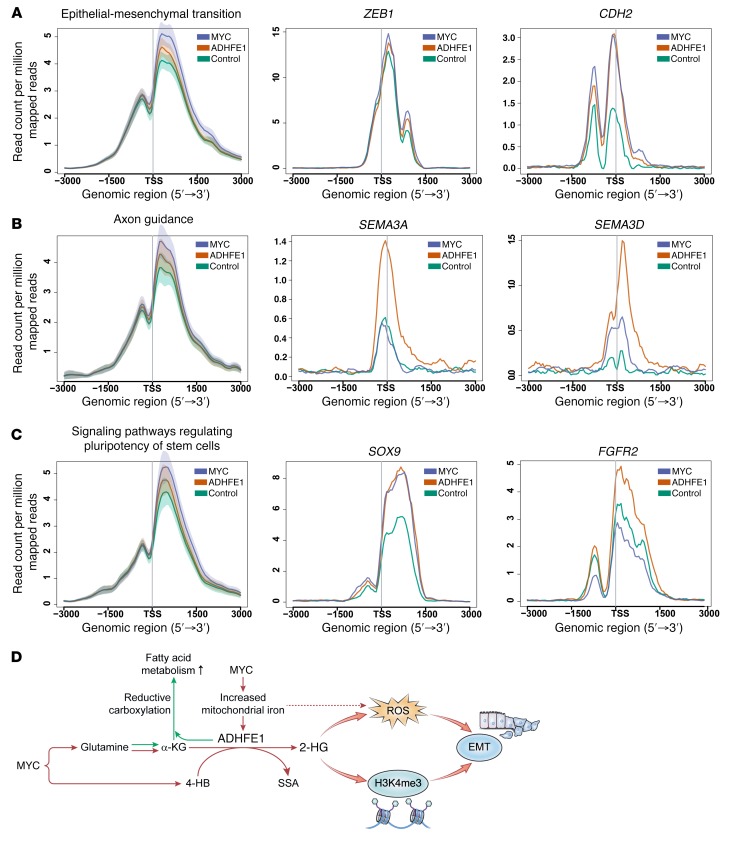
Figure 10. ChIP-seq analysis of H3K4me3 in ADHFE1- and MYC-overexpressing MCF7 cells.
Trimethylation of histone H3 lysine 4 (H3K4me3) is globally increased in promoter regions of genes regulating epithelial-to-mesenchymal transition (EMT) (A), the axon guidance pathway (B), and stemness (C) in MCF7 cells overexpressing either MYC or ADHFE1 (FDR < 5% for each pathway, compared with vector control cells). Examples for EMT pathway, ZEB1 and CDH2 (N-cadherin); axon guidance, SEMA3A and SEMA3D; and stem cell markers, SOX9 and FGFR2. H3K4me3 is significantly increased in the promoter region for each of the shown genes, with the exception of ZEB1, comparing ADHFE1-overexpressing versus vector control cells (FDR < 5%). EMT pathway: 344 genes. Axon guidance pathway: 127 genes. Pluripotency of stem cells pathway: 138 genes. Shown is the summarized read coverage for triplicate experiments of the ChIP-seq data. TSS, transcription start site. (D) Upregulation of ADHFE1 and D-2HG formation in breast tumors and their relationship to EMT and metabolic reprogramming. MYC increases D-2HG formation by increasing both substrate and iron availability for the iron-containing enzyme, ADHFE1. ADHFE1 and D-2HG induce a reductive glutamine metabolism, ROS, and histone methylation at H3K4me3, leading to increased acetyl-CoA and fatty acid synthesis and EMT. α-KG, α-ketoglutarate; 4-HB, 4-hydroxybutyrate; SSA, succinic semialdehyde.