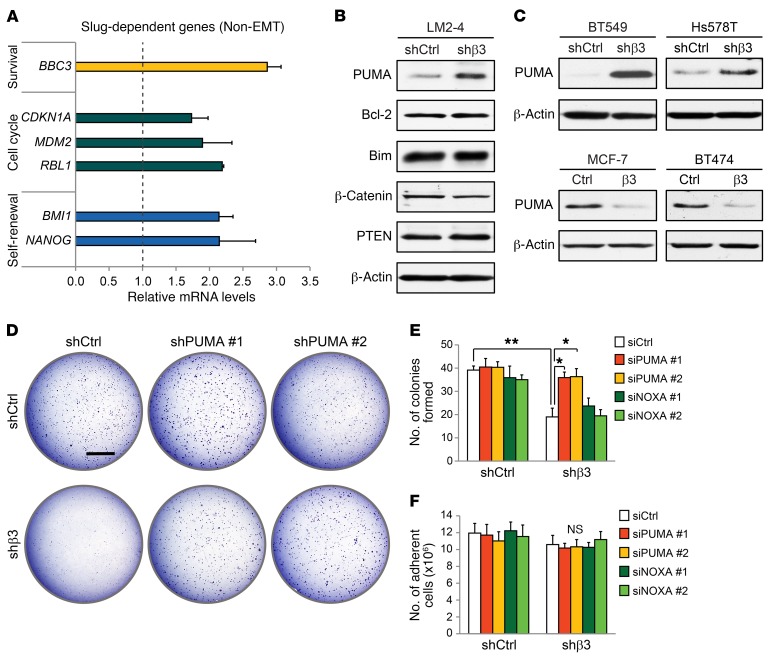
Figure 3. PUMA is a critical Slug-dependent gene suppressed by αvβ3 to promote anchorage independence.
(A) qPCR analysis of non-EMT, Slug-dependent genes in β3-knockdown LM2-4 cells. Fold change (2–ΔΔCT) in β3-knockdown cells is relative to cells expressing a control nonsilencing shRNA. Cyclophilin A was used as a loading control. (B and C) Immunoblots showing PUMA protein levels in breast cancer cell lines expressing either control nonsilencing shRNA or β3 shRNA (B and C, top) or ectopic β3 cDNA (β3) or a vector-only control (C, bottom). β-Actin was used as a loading control. (D) Soft agar tumorsphere assay comparing PUMA shRNA (shPUMA) knockdown in control and β3-knockdown LM2-4 cells. Scale bar: 2 mm. (B–D) Data shown are representative of 3 independent experiments. (E and F) Quantitation of tumorsphere formation per field (E) and total number of adherent cells (F) after siRNA knockdown of PUMA (siPUMA) or NOXA (siNOXA) compared with control siRNA (siCtrl) in LM2-4 cells, with or without β3. (E) P = 0.0072 (siCtrl; shCtrl vs. shβ3); P = 0.0331 (shβ3; siCtrl vs. siPUMA no. 1); P = 0.0275 (shβ3; siCtrl vs. siPUMA no. 2); (E and F) *P < 0.05 and **P < 0.01. Statistical analysis was performed by 2-way ANOVA with Tukey’s multiple comparisons test. Data represent the mean ± SEM. n = 3 independent experiments (A, E, and F). Each sample was run in triplicate. See also Supplemental Figure 4.