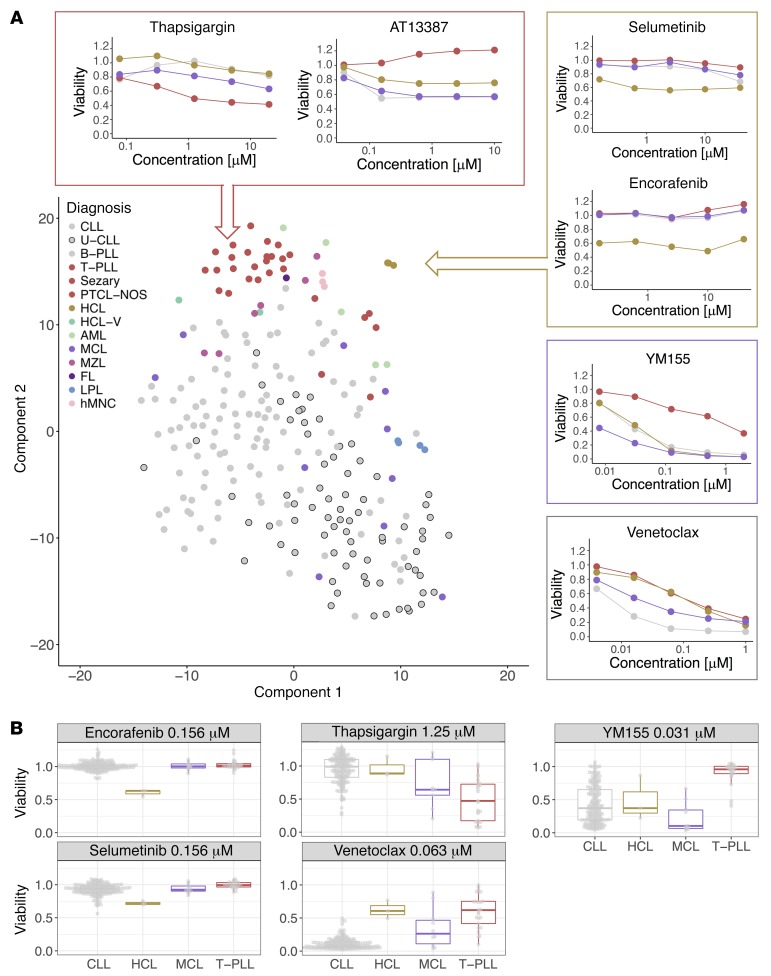
Figure 5. Disease-specific drug response phenotypes of blood cancers.
(A) t-distributed stochastic neighbor embedding (t-SNE), a machine learning algorithm for dimensionality reduction, was used to visualize similarities among 246 patient samples with respect to the 315 drug sensitivity measurements (each of 63 drugs at 5 concentrations). The plot shows a distinctive separation of pathologic disease entities based on their drug sensitivity pattern. The line plots show mean viabilities for individual disease entities (CLL, gray; HCL, yellow; MCL, purple; and T-PLL, brown) and drugs across 5 concentrations, highlighting disease-specific differences. (B) Primary data for individual drugs provide examples for disease-specific response and sample variation (CLL, n = 184; HCL, n = 3; MCL, n = 10; T-PLL, n = 25). FL, follicular lymphoma; HCL-V, hairy cell leukemia variant; hMNC, human mononuclear cell; LPL, lymphoplasmacytic lymphoma PTCL-NOS, peripheral T cell lymphoma not otherwise specified.