Abstract
Uncovering the causes of pregnancy complications such as preterm labor requires greater insight into how the uterus remains in a noncontractile state until term and then surmounts this state to enter labor. Here, we show that dynamic generation and erasure of the repressive histone modification tri-methyl histone H3 lysine 27 (H3K27me3) in decidual stromal cells dictate both elements of pregnancy success in mice. In early gestation, H3K27me3-induced transcriptional silencing of select gene targets ensured uterine quiescence by preventing the decidua from expressing parturition-inducing hormone receptors, manifesting type 1 immunity, and most unexpectedly, generating myofibroblasts and associated wound-healing responses. In late gestation, genome-wide H3K27 demethylation allowed for target gene upregulation, decidual activation, and labor entry. Pharmacological inhibition of H3K27 demethylation in late gestation not only prevented term parturition, but also inhibited delivery while maintaining pup viability in a noninflammatory model of preterm parturition. Immunofluorescence analysis of human specimens suggested that similar regulatory events might occur in the human decidua. Together, these results reveal the centrality of regulated gene silencing in the uterine adaptation to pregnancy and suggest new areas in the study and treatment of pregnancy disorders.
Keywords: Reproductive Biology
Keywords: Epigenetics, Fibrosis, Obstetrics/gynecology
Introduction
The decidua, a tissue derived from the endometrium and anatomically positioned between the conceptus and the myometrium, has long been thought to play a central role in pregnancy. For most of gestation, it assumes a relatively “quiescent” state while providing trophic and structural support to the placenta (1). At term, it enters an activated state that, through the production of inflammatory mediators, is thought to promote myometrial contractility and labor induction (2, 3). The exact natures and molecular determinants of these 2 states remain largely undefined, however, which precludes a clear understanding of how the pregnant uterus transitions from quiescence to labor. This question has considerable clinical importance given the high and increasing world-wide incidence of preterm labor, a major cause of neonatal mortality and morbidity that has no known etiology in the absence of infection (4, 5). Since women with idiopathic preterm labor have normal serum levels of the quintessential pregnancy hormone progesterone (P4), it has been suggested that the disorder, at least in some cases, results from a “functional” P4 withdrawal that renders the uterus resistant to P4 levels otherwise sufficient to sustain pregnancy (6). The mechanistic basis of this process, however, remains undefined.
Previously, we found in mice that the chemokine genes Cxcl9 and Cxcl10 are transcriptionally silenced in decidual stromal cells (DSCs), the fibroblastic constituents of the decidua and descendants of endometrial stromal cells (ESCs) (7). This occurred in association with promoter accrual of tri-methyl histone H3 lysine 27 (H3K27me3), a repressive histone mark generated by polycomb repressive complex 2 (PRC2), and resulted in exclusion of effector Th1 cells and CD8+ cytotoxic T lymphocytes (CTLs) from the maternal-fetal interface. These results revealed a new function for the decidua — protecting the conceptus from attack by maternal T cells — as well as the underlying molecular mechanism. Here, we show that H3K27me3 dynamics in DSCs both reveal and explain aspects of decidual biology with potentially greater significance, with H3K27me3 deposition in early gestation actively preventing the decidua from mounting tissue reactions that might trigger premature uterine contraction, and relief from this program via scheduled H3K27 demethylation in late gestation conversely driving decidual activation and labor entry.
Results
H3K27me3 accrues on a broad set of target genes in early gestation DSCs.
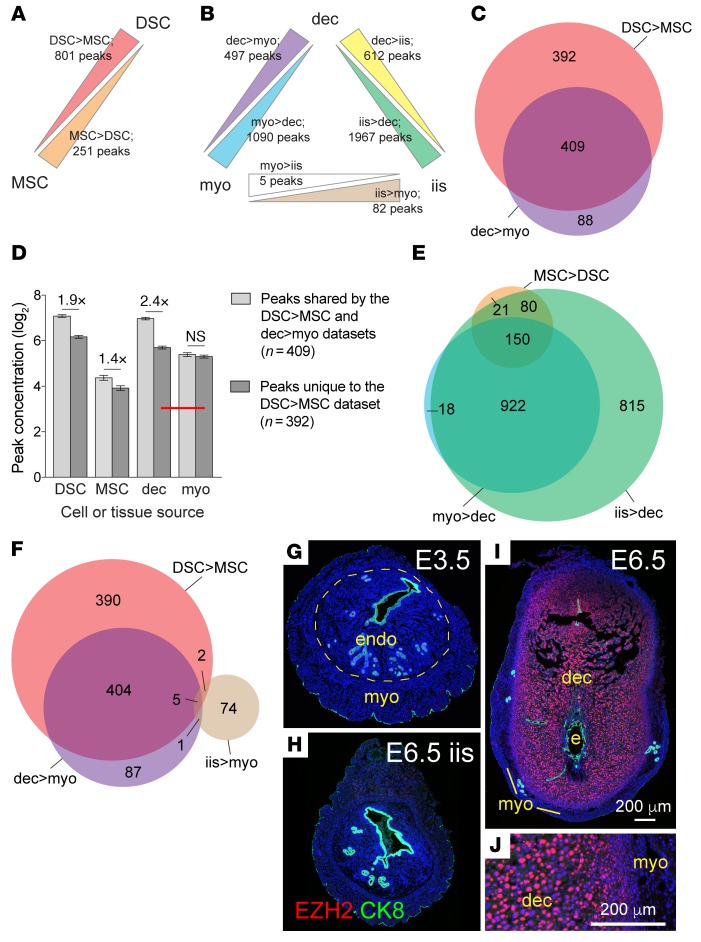
To identify all protein-coding genes enriched for H3K27me3 in DSCs, we subjected purified early gestation DSCs and companion myometrial stromal cells (MSCs) to H3K27me3 ChIP followed by deep sequencing (ChIP-Seq). We identified 801 peaks that were enriched more than 2-fold in DSCs (termed DSC>MSC peaks), corresponding to 822 genes, and 251 peaks that were enriched more than 2-fold in MSCs (MSC>DSC peaks), corresponding to 257 genes (Figure 1A, Supplemental Figure 1, A–D, and Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI95937DS1). Importantly, 51% of the DSC>MSC peaks (409/801) were also present in the set of peaks found by ChIP-Seq to be enriched in extracts of whole (i.e., unfractionated) E7.5 decidua as compared with whole, overlying myometrium (collectively termed dec>myo peaks), confirming their presence in early gestation implantation sites in vivo (Figure 1, B and C, and Supplemental Tables 1 and 2). This 51% was, moreover, likely an underestimation, since some DSC>MSC peaks were probably undetectable as differentially enriched between whole-tissue layers due to the compounded effects of their low concentration and interference from the nonstromal cell components of whole-tissue myometrial extracts (Figure 1D).
Figure 1. H3K27me3 generation in the postimplantation uterus.
(A and B) Schematics of ChIP-Seq comparisons. We considered only peaks that overlapped or were less than 5 kb upstream of the TSS (4,504 total). Stromal cells (DSCs, MSCs) were cultured for 24 hours after purification from artificially decidualized uteri (an abundant source of cells) on the day corresponding to E7.5. Comparisons between these cells (A; n = 3 each) tallied peaks that were significantly enriched more than 2-fold between populations (FDR < 0.05). Comparisons between whole tissues and tissue layers (B; n = 3 decidua and interimplantation site samples, n = 2 myometrium samples, all from true pregnancies on E7.5) employed the same overall peak set but, in consideration of the nonstromal cell contributions to the extracts, tallied those peaks with significantly different concentrations regardless of the degree of difference. (C, E, and F) Venn diagrams indicating the overlap between various peak sets. (D) Absolute concentrations of DSC>MSC peaks in stromal cell and whole-tissue layer extracts (mean ± SEM). Significant differences are indicated (unpaired t test). The lower concentration of peaks unique to the DSC>MSC data set brought their average concentration in decidual extracts into proximity with their average concentration in the whole-tissue myometrial extracts (red bar), which was not reduced in parallel. This precluded their scoring as enriched between tissue layers. Similar considerations applied to MSC>DSC peaks, which nonetheless already showed 60% overlap with myo>dec peaks (E, Supplemental Figure 1C). (G–J) EZH2 immunostaining of peri-implantation uteri. On E3.5 and in E6.5 interimplantation sites, EZH2 was expressed at low or undetectable levels by ESCs (G and H), whereas on E5.5 (not shown) and E6.5 (I), it was strongly expressed in most DSCs. Panels G–I are shown at the same magnification. Panel J is a close-up of panel I; note the nuclear accumulation of EZH2 in a large fraction of DSCs. CK8 identifies uterine epithelial cells and trophoblasts. DAPI counterstain. myo, myometrium; endo, endometrium; dec, decidua; e, embryo.
Next, we performed ChIP-Seq on whole-tissue extracts prepared from the segments of undecidualized uterus between implantation sites (henceforth referred to as interimplantation sites) on E7.5 in order to gauge the extent to which DSC>MSC peaks were enriched for H3K27me3 in ESCs prior to decidualization (Figure 1B). This question could not be answered through direct analysis of ESCs given the technical limitations on isolating these cells as pure populations. Strikingly, 92% of the MSC>DSC peaks and 98% of the peaks enriched in whole-tissue extracts of myometrium compared with decidua (myo>dec peaks) were present in the set of peaks enriched in whole-tissue extracts of interimplantation sites compared with decidua (iis>dec peaks), but only 0.9% of the DSC>MSC peaks and 1.2% of the dec>myo peaks were present in the set of peaks enriched in whole-tissue extracts of interimplantation sites compared with myometrium (iis>myo peaks) (Figure 1, E and F, and Supplemental Table 2). In other words, interimplantation sites, comprising undecidualized endometrium and myometrium, contained myometrium-specific peaks, but not decidua-specific peaks. These results indicated that DSCs accrued H3K27me3 on target genes de novo during their implantation-induced transformation from ESCs. In further support of this conclusion, enhancer of zeste homolog 2 (EZH2), the catalytic subunit of PRC2, was highly but transiently induced within DSCs of early implantation sites (Figure 1, G–J, and Supplemental Figure 1, E–I).
H3K27me3-induced gene silencing in DSCs reveals and explains aspects of decidual biology.
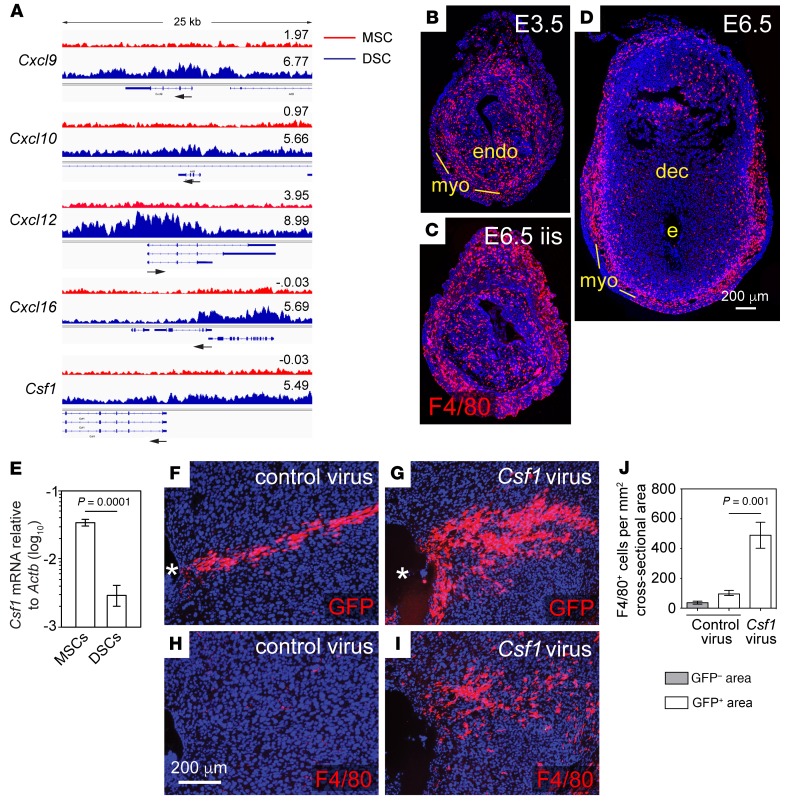
DSC>MSC H3K27me3 target genes included Cxcl9 and Cxcl10, confirming our previous results (7), as well as Cxcl12 and Cxcl16 (Figure 2A). Aside from Ccl5, which lacked H3K27me3 but was nonetheless silenced in DSCs (see below), these genes together encode the complete set of chemokines capable of recruiting Th1 cells and CTLs to peripheral tissues (8). The only other chemokine gene with enriched H3K27me3 in DSCs was Cxcl5. DSC>MSC H3K27me3 target genes also included Csf1, encoding the prototypical macrophage growth factor CSF-1 (Figure 2A), which explained previous observations that the decidua expresses low levels of Csf1 and contains few macrophages, even following infection (9, 10) (see also Figure 2, B–E). Indeed, lentivirus-mediated CSF-1 expression within the decidua induced focal macrophage accumulation, formally demonstrating that the decidua’s CSF-1 deficit was functional (Figure 2, F–J). Since macrophages both amplify and serve as effectors in type 1 immune responses mediated by Th1 cells and CTLs, these results together revealed that the suppression of type 1 immunity within the decidua was one general role for H3K27me3 accrual in DSCs. Notably, H3K27me3 did not generally suppress the inflammatory responses of DSCs, with genes such as Stat1, Gbp5, Gbp7, Saa3, and Irf1 being highly inducible in DSCs following combined treatment with TNF-α and IFN-γ; conversely, certain inflammatory targets, such as Ccl5, were silenced in DSCs yet lacked H3K27me3 enrichment (Supplemental Figure 2).
Figure 2. H3K27me3 accrual in DSCs attenuates type 1 immunity in the decidua.
(A) H3K27me3 tracks at the Cxcl9, Cxcl10, Cxcl12, Cxcl16, and Csf1 loci. The tracks are the pileups of the 3 independent replicates. The log2 concentration for each called peak and the TSS with direction of transcription (arrow) are indicated. Sequencing reads from total input chromatin were homogeneous across these loci (not shown). (B–J) H3K27me3 silences Csf1 in DSCs, limiting macrophage accumulation in the decidua. (B–D) Immunostaining for F4/80+ macrophages (red) in the undecidualized E3.5 uterus 1 day prior to implantation (B), an interimplantation site on E6.5 (C), and an E6.5 implantation site (D). Note the dramatically lower tissue density of macrophages within the decidua as compared with the undecidualized endometrium, consistent with previous results at later gestation (9). Representative images from 3 mice/group; panels C and D show sections near those in Figure 1, H and I, respectively. (E) qRT-PCR determination of Csf1 mRNA expression in stromal cells isolated from E7.5 pregnant mice and cultured for 24 hours (mean ± SEM; n = 3 samples/group). DSCs express lower levels of Csf1 than MSCs, at least in part explaining the lower level of Csf1 expression in the whole decidua (9). (F–J) Effect of ectopic CSF-1 expression within the decidua. Artificially decidualized uteri were injected with Csf1-expressing or empty vector control lentivirus on the day corresponding to E5.5 and the mice were sacrificed 2 days later. Viral preparations included aliquots of EGFP reporter lentiviruses to identify transduced areas. Representative images of serial sections immunostained for F4/80 or GFP (F–I) and mean ± SEM of quantified F4/80+ cell densities in infected (GFP+) and uninfected (GFP–) decidual areas (J) from 4 control virus–infected mice and 5 Csf1 virus–infected mice. Note that decidual areas infected with Csf1-expressing lentiviruses accumulate macrophages, demonstrating a CSF-1 deficit within this tissue layer. Asterisk indicates decidual lumen.
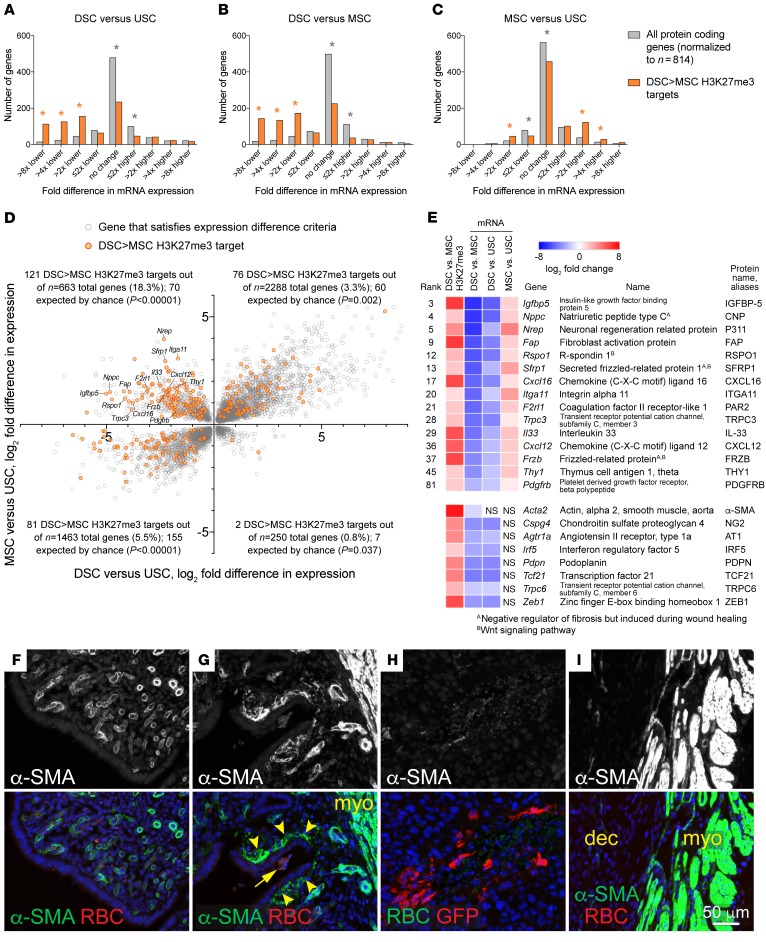
To identify additional roles for H3K27me3 accrual in DSCs, we cross-correlated our ChIP-Seq analysis with an RNA-sequencing (RNA-Seq) analysis of E7.5 DSCs, MSCs, and the stromal cells isolated from the uteri of P4-treated virgin mice (Supplemental Table 3). Consisting of roughly equal proportions of ESCs and the stromal cells of the overlying (nongrowing) myometrium (11), this latter population (termed uterine stromal cells [USCs]) could be isolated in abundance and served as surrogates for the stromal cells comprising interimplantation sites, thus providing a baseline for uterine gene expression as influenced by P4, but independent of the formation and growth of implantation sites. As expected, DSC>MSC targets were expressed at much lower levels in DSCs as compared with both USCs and MSCs, indicating that transcriptional silencing was the general outcome of H3K27me3 target gene accrual in DSCs (Figure 3, A and B). However, DSC>MSC targets were also expressed at higher levels in MSCs compared with USCs (Figure 3C) and, moreover, were greatly enriched within the set of genes with the dual characteristics of increased expression in MSCs compared with USCs, but decreased expression in DSCs compared with USCs (Figure 3D and Supplemental Table 4). When these targets were ranked based upon how much their expression ultimately diverged between DSCs and MSCs, 33% of the top 50 were noted to encode markers or positive regulators of fibroblast activation, myofibroblast formation, or fibrosis, related processes induced during wound healing and seen in reactive cancer stroma (12–14) (Figure 3E; H3K27me3 tracks for these and other relevant genes are shown in Supplemental Figure 3). The list of all DSC>MSC targets included even more of these markers, including the signature myofibroblast marker smooth muscle actin (α-SMA, also called ACTA2) (Figure 3E). These results suggested that the decidua had the previously unappreciated property of suppressing fibroblast activation.
Figure 3. H3K27me3 accrual in DSCs prevents fibroblast activation/myofibroblast formation and wound healing in the decidua.
(A–C) RNA-Seq analysis of DSCs, MSCs, and USCs, and the relationship of gene expression to H3K27me3 marking. Distribution of mRNA expression differences (Padj < 0.05; n = 3 samples/group), binned into progressive 2-fold changes. *P < 5.6 × 10–3, significantly over- (orange) or underrepresented (gray) for DSC>MSC targets as compared with expectation. (D) DSC>MSC H3K27me3 targets within all genes with significantly different expression both between MSCs and USCs and between DSCs and USCs. (E) H3K27me3 marking and mRNA expression of DSC>MSC targets involved in fibroblast activation/myofibroblast formation. The 15 genes with rankings are from the upper left quadrant of D and encode the classical markers FAP (12), CXCL12 (12), THY1 (14), and PDGFRB (12), proteins with less extensive literature such as IGFBP-5 (45), C-type natriuretic peptide (46), P311 (13), CXCL16 (47), integrin α11 (13), PAR2 (48), TRPC3 (49), and IL-33 (50), and several Wnt-signaling regulators induced during fibrosis or themselves profibrotic (13, 51). Although their expression was not increased in MSCs compared with USCs, other DSC>MSC targets included Acta2 (α-SMA) (13) and Cspg6 (14), 2 other major myofibroblast markers, as well as Agtr1a (14), Irf5, Pdpn (12), Tcf21 (14), Trpc6 (14), and Zeb1 (12), encoding additional emerging markers or inducers (bottom set). (F–I) Impaired wound healing responses in the decidua. Paired horns of undecidualized uteri were left unmanipulated (F) or scratched along their inner surface with a needle (G). Note the autofluorescent RBCs at the wound site (arrow). Representative images from 5 mice. Artificial decidua were wounded via injection with EGFP-expressing lentiviruses. Note the extravascular RBCs but minimal α-SMA staining at an injection site (H); the nearby myometrium was α-SMA+ (I). Representative images from 52 injection sites from 4 mice. Mice were sacrificed 2 days after scratching/injection.
We next tested whether such suppression translated into impaired wound healing within the decidua. As expected, scratching the epithelial surface of the undecidualized endometrium induced the formation of subepithelial α-SMA+ myofibroblasts at the wound site (Figure 3G, arrowheads), whereas such cells did not exist in the unscratched endometrium (Figure 3F). In contrast, when artificial decidua were injected with EGFP-expressing lentiviruses, which allowed the ensuing wounds to be identified by anti-GFP immunostaining, the wound sites displayed extravascular RBCs but only minimal α-SMA staining (Figure 3H). The nearby myometrium in these sections was α-SMA+, as expected (Figure 3I). Thus, the murine decidua is impaired in its ability to generate myofibroblasts and mount typical wound-healing responses.
Advancing gestational age is associated with genome-wide H3K27 demethylation in DSCs and target gene upregulation.
Unexpectedly, H3K27me3 accrual in DSCs also silenced Ptgfr and Oxtr, which encode the prostaglandin PGF2α receptor (FP) and oxytocin receptor (OXTR) (6.4- and 10.8-fold lower expression in DSCs versus USCs, respectively; Supplemental Figure 3B and Supplemental Tables 1 and 3). This finding revealed an additional, directly anticontractile function for the gene-silencing program, since PGF2α and oxytocin are the primary hormones that drive uterine contraction during labor (3, 15). On the other hand, the finding was paradoxical given that the functionality of PGF2α and oxytocin at term is thought in part to require Ptgfr and Oxtr upregulation in the decidua. Indeed, such upregulation is one of the hallmarks of decidual activation (2, 16).
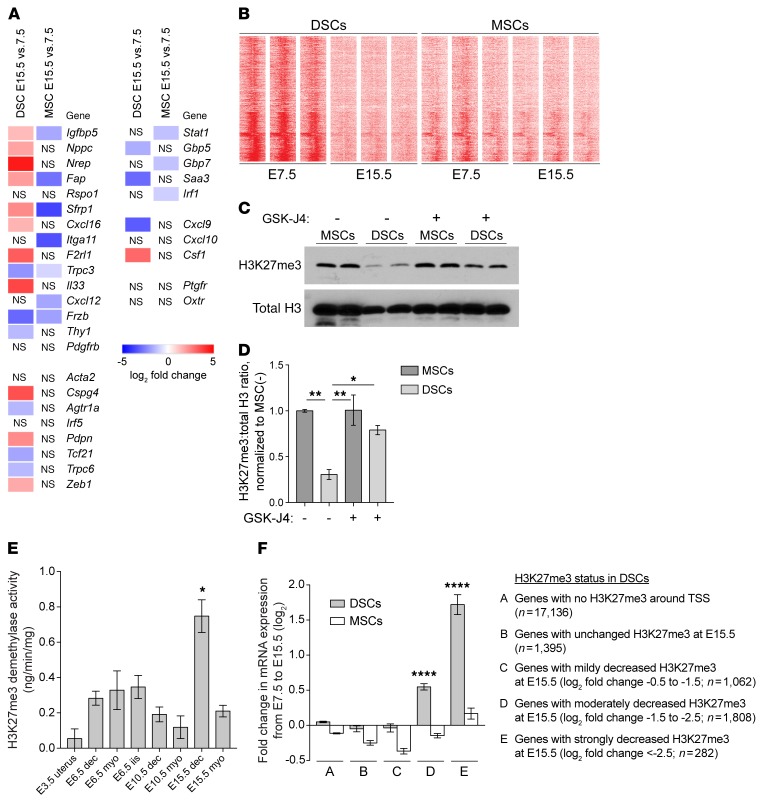
Also unexpectedly, an RNA-Seq analysis of stromal cells isolated from implantation sites in late gestation (i.e., E15.5, which is 3 days earlier than term [E18.5] for the B6CBAF1 mice used in this study) revealed that 33% of the H3K27me3 DSC>MSC targets (268/814) were significantly (Padj < 0.05) upregulated in DSCs when compared with E7.5 while 21% (174/814) were downregulated, a general trend toward upregulation that was not evident in MSCs (141 up and 211 down; P = 0.003 compared with DSCs, Fisher’s exact test) (Supplemental Table 5). With respect to the 15 H3K27me3-silenced fibroblast activation markers listed in the top half of Figure 3E, 8 were upregulated in DSCs, but none of them were upregulated in MSCs (P = 0.002, Fisher’s exact test) (Figure 4A). A similar pattern extended to other fibroblast activation markers with elevated H3K27me3 in DSCs (bottom portions of Figure 3E and Figure 4A) and was also evident from a quantitative reverse-transcriptase PCR (qRT-PCR) analysis of whole-tissue layers, which revealed that decidual expression of Nppc, Nrep, Fap, Cxcl16, Itga11, F2rl1, Il33, Cxcl12, and Pdgfrb increased with advancing gestation to approach or even exceed expression in the myometrium by E17.5 (Supplemental Figure 4A). We documented upregulated protein expression of 2 of these markers, Fap and F2rl1, by immunofluorescence (Supplemental Figure 5). Provocatively, Stat1, Gbp5, Gbp7, Saa3, and Irf1 — i.e., genes we had found to be highly inducible in DSCs following combined TNF-α/IFN-γ stimulation (Supplemental Figure 2B) — did not show a parallel pattern of upregulation, despite the prevailing idea that decidual activation is an inflammatory process (2, 16). Rather, the expression of these inflammatory targets either decreased or held relatively constant up through E17.5 (Figure 4A, and Supplemental Figure 4D, which includes additional analysis). With the exception of Saa3, whose H3K27me3 levels were similar between DSCs and MSCs on E7.5, none of these inflammatory targets had detectable H3K27me3 in DSCs or MSCs (Supplemental Figure 3C).
Figure 4. H3K27 demethylation in late-gestation DSCs, associated with gene upregulation.
RNA-Seq (A and F) and ChIP-Seq (B and F) were performed on purified cells cultured for 24 hours (n = 3 samples/group). (A) RNA-Seq analysis of various H3K27me3 DSC>MSC targets and inducible inflammatory targets. Other genes of interest are also shown. (B) Distribution of H3K27me3 ±5 kb around the TSS of DSC>MSC targets. Replicates are shown. (C and D) Representative Western blots (C) and normalized H3K27me3:total H3 levels (D) of cells isolated from E15.5 mice injected daily with GSK-J4 or vehicle (–) from E13.5 and cultured for 24 hours (mean ± SEM of n = 4 samples/group; 2 sets of blots). For each set, the average H3K27me3/total H3 ratio for the MSC samples from vehicle-treated mice was set to 1.0 (P = 0.0004, 1-way ANOVA; *P = 0.012; **P < 0.001). Of note, E15.5 DSCs contained less total H3 than MSCs; GSK-J4 had no effect on levels of H3K9me3, another repressive histone mark (Supplemental Figure 6, C–G). (E) H3K27me3 demethylase activity measured on nuclear extracts of whole tissue or tissue layers (mean ±S EM; n = 3 mice/group). The spike in activity in the E15.5 decidua was significantly different from all other groups (P < 0.0001, 1-way ANOVA; *P < 0.05 compared with all other groups). (F) Relationship between H3K27me3 status in DSCs and gene-expression changes (mean ± SEM; n = 3 samples/group). Significantly different DSC groups are indicated (P < 0.0001, 1-way ANOVA; ****P < 0.0001 compared with all other DSC groups). Genes with undetectable H3K27me3 on E7.5 showed no average expression change from E7.5 to E15.5, as did genes for which H3K27me3, while detectable, was either insignificantly changed or reduced less than 1.5 log2–fold. In contrast, greater degrees of H3K27me3 loss correlated with increasingly greater degrees of upregulation. This pattern was not evident for MSCs. Each DSC group was significantly different from its respective MSC group (P < 0.0001, paired t test).
Together, these results suggest that a gene’s expression level in late gestation was determined by its H3K27me3 status, potentially independently of inflammation. To examine this possibility more directly, we performed ChIP-Seq on E15.5 DSCs and MSCs. Strikingly, 84% of the DSC>MSC peaks (674/801) showed significantly reduced concentrations in DSCs on E15.5 compared with E7.5, whereas H3K27me3 profiles were virtually unchanged in MSCs (Figure 4B and Supplemental Table 6). H3K27me3 loss was nonuniform across the DSC genome, which strongly argued against technical artifacts in the ChIP-Seq experiment (e.g., low amounts of input chromatin; Supplemental Figure 6, A and B). It was, moreover, evident by Western blot and could be specifically inhibited by administration of GSK-J4, an inhibitor of the 2 known H3K27me3 demethylases KDM6A and KDM6B (17) (Figure 4, C and D, and Supplemental Figure 6, C and D). Indeed, H3K27me3 demethylase enzyme assays performed on nuclear extracts from E3.5, E6.5, E10.5, and E15.5 uterine tissue layers revealed a spike in activity in the E15.5 decidua (Figure 4E). This contrasted with the low level of EZH2 expression in late gestation, but coincided with H3K27me3 loss from DSCs, which became evident after E12.5 (Supplemental Figure 1, F–I, and Supplemental Figure 6, E–G). The degree of H3K27me3 loss from genes in E15.5 DSCs, moreover, predicted their level of transcriptional upregulation (Figure 4F and Supplemental Table 5). Together, these results suggested that advancing gestation was associated with active H3K27 demethylation in DSCs and upregulation of the demethylated genes.
Of interest, the induction of H3K27me3 demethylase activity in the E15.5 decidua did not obviously correlate with transcriptional induction of Kdm6a or Kdm6b, which, respectively, encode KDM6A and KDM6B. The levels of these transcripts in the decidua remained relatively constant over the course of gestation and were only increased on E17.5 (Supplemental Figure 4E). Moreover, Kdm6a was induced only modestly and transcript levels of this gene were generally similar in the decidua and myometrium. Levels of PRC2 component–encoding transcripts also remained relatively constant over the course of gestation and did not dramatically differ between decidua and myometrium (Supplemental Figure 4E), despite the transient appearance of EZH2 protein and the de novo induction of H3K27me3 marks specifically in early gestation DSCs. Our RNA-Seq analysis of DSCs and MSCs also failed to reveal tissue layer– and gestation stage–specific differences in gene expression that would easily explain ultimate enzyme activities (Supplemental Table 5). These observations suggest that H3K27 methylase and demethylase activities are induced in early and late-gestation DSCs, respectively, largely through posttranscriptional mechanisms, including potentially regulated protein stability.
KDM6A/B inhibition prevents decidual activation and labor induction.
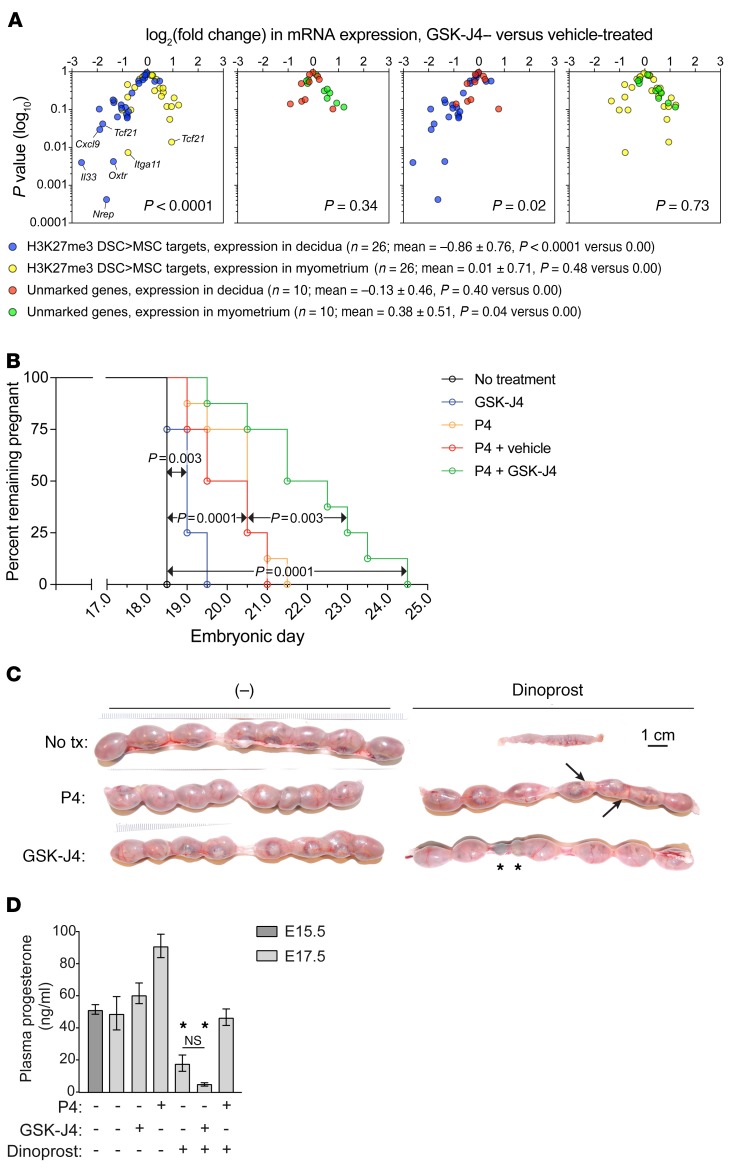
We next determined the effect of GSK-J4 on gene expression in the late-gestation uterus. Although rarely reaching the P < 0.05 level of significance for individual genes (which is not surprising given that the qRT-PCR assays were performed on RNA isolated from whole-tissue layers), GSK-J4 treatment reduced the aggregate expression of H3K27me3 targets in the decidua, but not the myometrium (Figure 5A, blue and yellow, and Supplemental Figure 7, A and B). This could not be ascribed to altered mRNA expression of genes encoding PRC2 components or H3K27me3 demethylases, nor did GSK-J4 significantly affect mRNA expression of Pgr (which encodes the P4 receptor) or tissue layer–specific markers (Figure 5A, red and green, and Supplemental Figure 7C). Indeed, given that these latter genes also all lacked detectable H3K27me3, we used them as an internal control group to discriminate the effects of GSK-J4 that were H3K27me3 dependent versus independent. Accordingly, we found that GSK-J4 decreased the expression of H3K27me3 targets relative to this control group in the decidua (Figure 5A, blue and red), but had no differential effect on the 2 groups in the myometrium (Figure 5A, green and yellow). Together, these observations suggested that the effects of GSK-J4 on uterine gene expression were inhibitory, largely specific to H3K27me3-marked genes, and largely confined to the decidua.
Figure 5. Effect of GSK-J4 on late-gestation pregnancy and parturition.
Mice were injected daily with GSK-J4 or vehicle starting on E13.5. (A) qRT-PCR analysis of mRNA expression in dissected decidual and myometrial tissue layers on E17.5, with log2(fold change) between GSK-J4– and vehicle-treated mice displayed on volcano plots (n = 6 mice/group). Genes affected by GSK-J4 (P < 0.05) are indicated. Supplemental Figure 7, A–C, shows individual gene-expression levels; the unmarked genes encode epigenetic modifiers, tissue layer–specific markers, and Pgr. The symbol key lists mean log2(fold change) ± SD for each group and the P value of its comparison to 0.00 by 1-sample t test. Groups were compared with each other by 1-way ANOVA (P < 0.0001) followed by Sidak’s multiple comparison test (P values shown in each plot). (B) Effects of GSK-J4 and P4 on delivery time (n = 8 mice/group); P values were determined by the log-rank (Mantel-Cox) test. (C) Effect of GSK-J4 on dinoprost-induced preterm delivery. Mice were injected with dinoprost and/or P4 on E16.5. Representative images from 6–8 mice/group. Injection of E16.5 pregnant mice with dinoprost unfailingly induced delivery within 24 hours (0/7 mice remained pregnant), 1 day before term. None of the delivered pups were viable. While P4 cotreatment prevented early delivery in all cases (6/6), only 27% of the fetuses were viable. Arrows indicate 2 pale, dead fetuses. In contrast, GSK-J4 cotreatment both prevented delivery (7/7 mice pregnant; P = 0.0006 compared with dinoprost alone, Fisher’s exact test) and maintained pup viability (86%; P < 0.0001 compared with dinoprost plus P4, Fisher’s exact test). Asterisks indicate implantation sites that had resorbed earlier in gestation. (D) Plasma P4 levels (mean ± SEM; n = 3 mice/group). GSK-J4/dinoprost–treated mice had low plasma P4 (P < 0.0001, 1-way ANOVA; *P < 0.0001) and thus had undergone luteolysis; presumably, they would soon enter labor. Supplemental Table 7 shows a full accounting of the experiments in panels B and C.
We next determined how GSK-J4 affected the timing of normal labor. Importantly, GSK-J4 did not inhibit expression of 20α-hydroxysteroid dehydrogenase, the P4-catabolizing enzyme induced at term in the mouse ovary through the process of luteolysis (Supplemental Figure 7D), but nevertheless delayed delivery for about 24 hours (Figure 5B, 98% pup viability; Supplemental Table 7). Conversely, P4 administration, with or without the vehicle for GSK-J4, delayed parturition 2 to 3 days (40%–50% pup viability), consistent with prior results and the idea that intrauterine processes are sufficient to drive labor induction in the presence of P4 (3, 18). Delivery was strikingly delayed, however, in mice given both GSK-J4 and P4, with those still pregnant after E21.5 needing to be sacrificed due to maternal distress (0% pup viability). Together, these data suggested that inhibition of decidual H3K27 demethylation prevents spontaneous labor onset when the decline in ovarian function that occurs at the end of mouse gestation is rescued by exogenous P4 injection.
We also determined whether GSK-J4 could inhibit the induction of preterm labor. One preterm labor model in mice is LPS injection, which is thought to recreate the intrauterine inflammatory state seen with acute chorioamnionitis (19). However, this model has been criticized because LPS injection also induces systemic inflammation and thus has multiple nonuterine effects (6). Indeed, GSK-J4 did not prevent LPS-induced preterm labor (data not shown). Thus, we developed a preterm parturition model based upon the use of dinoprost, a synthetic PGF2α used to provoke labor in the clinic due to its procontractile effect on the uterus (15). Importantly, dinoprost induced preterm delivery within 24 hours when given on E16.5 (Figure 5C), but unlike LPS, did not induce systemic inflammation (Supplemental Figure 7E) or cause the mice to appear acutely ill (data not shown). Strikingly, GSK-J4 prevented dinoprost-induced delivery and maintained pup viability (Figure 5D). The effect was not due to an inhibition of luteolysis, another effect of PGF2α in mice (15), and contrasted with that of exogenous P4, which prevented PGF2α-induced delivery but, consistent with unopposed dinoprost-induced uterine contractions, not fetal demise (Figure 5, C and D). Together, these data suggest that inhibition of decidual H3K27 demethylation maintains uterine quiescence and prevents both spontaneous and noninflammatory preterm birth.
Fibroblast activation marker and KDM6A expression in the pregnant human uterus.
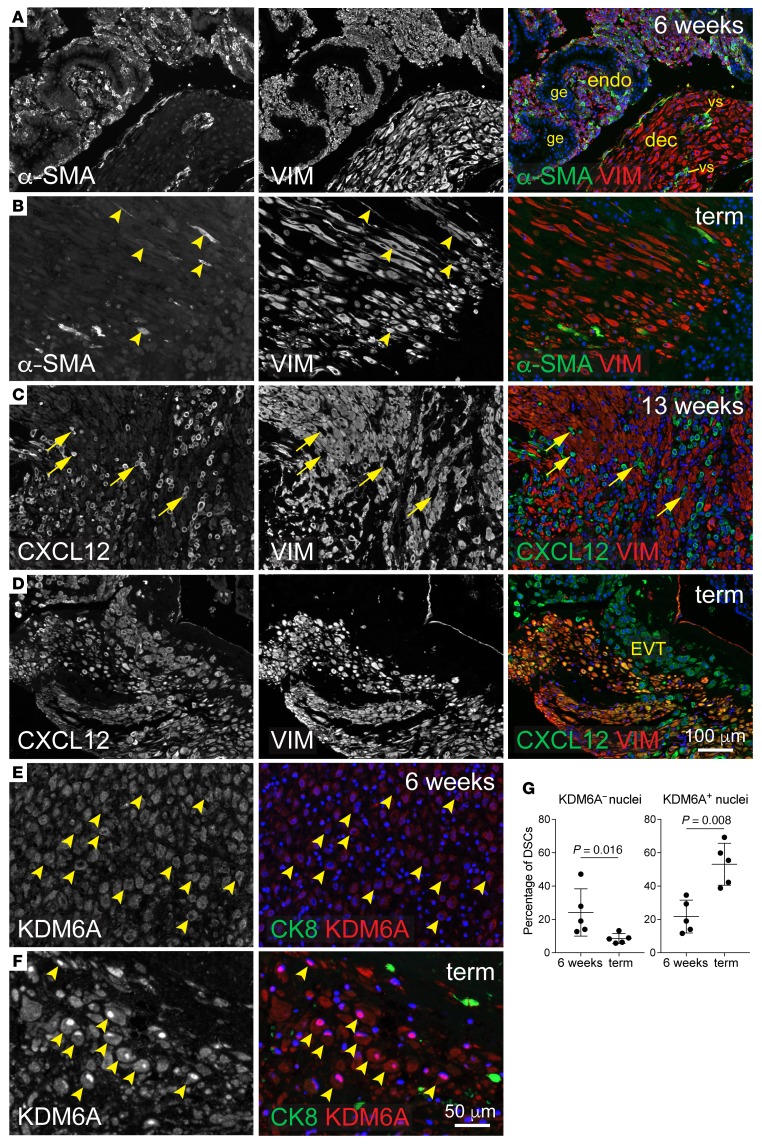
Finally, we sought to determine whether the pattern of transiently suppressed gene expression in DSCs found parallels in human pregnancy. At 6 and 13 weeks of gestation, uterine tissues from elective pregnancy termination specimens that bore no evidence of placental or decidual pathology (inflammatory or otherwise) revealed α-SMA expression by ESCs in areas of undecidualized endometrium, consistent with previous work on the nonpregnant uterus (20) (Figure 6A and data not shown). This suggests that human ESCs, at baseline, have myofibroblast-like characteristics. Strikingly, however, DSCs within decidualized areas did not express α-SMA, even at sites of trophoblast invasion (Figure 6A and Supplemental Figure 8, A and B). Thus, α-SMA, which is silenced by H3K27me3 in mouse DCSs, is downregulated in human DSCs in vivo. Extending the mouse-human parallel to late gestation, placental specimens from nonlaboring women at term contained scattered α-SMA+ DSCs within the basal plate, suggesting α-SMA reexpression (Figure 6B, arrowheads, and Supplemental Figure 8C), while CXCL12 (also called stromal cell derived factor-1 [SDF-1]) and platelet-derived growth factor receptor β (PDGFRB), 2 other classic fibroblast activation markers also silenced in mouse DSCs (Figure 3E), were not expressed at 6 or 13 weeks in either ESCs or DSCs, but were uniformly expressed by term DSCs (Figure 6, C and D, Supplemental Figure 8, D and E, and Supplemental Figure 9, A and B). These term specimens were obtained by elective Caesarian section and again were screened to confirm the absence of any placental or decidual pathology, inflammatory or otherwise. A third marker, chondroitin sulfate proteoglycan 4 (CSPG4), was undetectable at all time points (Supplemental Figure 9C and data not shown). Strikingly, KDM6A showed greater nuclear accumulation at term compared with 6 weeks, suggesting increased activity in late gestation (Figure 6, E–G). Thus, with variation between species at the level of individual markers and exact expression patterns, these observations suggest that mouse and human DSCs shared the common features of suppressed fibroblast activation in early gestation yet manifested activated fibroblast/myofibroblast phenotypes in late gestation in association with KDM6A induction.
Figure 6. Expression of fibroblast activation markers and KDM6A in the pregnant human uterus.
Images are representative of 5 specimens per time point. Specimens at 6 and 13 weeks were from elective terminations. Vimentin (VIM) was used as an ESC/DSC marker. DAPI counterstain (blue). (A and B) α-SMA/VIM staining. At 6 weeks (A), fields of small VIM+ cells associated with extensive glandular epithelium (ge) identify areas of undecidualized endometrium. α-SMA+ cells within decidualized areas at 6 (A) and 13 weeks (Supplemental Figure 8B) were invariably associated with vascular structures (vs) and thus were vascular smooth muscle cells. (C and D) CXCL12/VIM staining. Note the absence of CXCL12 expression in DSCs at 13 weeks, which contrasts with interspersed CXCL12+VIM– cells (C, arrows). Neighboring sections revealed these VIM– cells to be positive for CK7 and CK8 (not shown and Supplemental Figure 8A), identifying them as extravillous trophoblasts (EVT) and confirming their expression of CXCL12 described previously (52). At term, all DSCs expressed substantial CXCL12 levels that were similar to those of neighboring trophoblasts (D). (E and F) KDM6A/CK8 staining; higher magnification. At 6 weeks, KDM6A staining in DSCs (identified morphologically and by their CK8 negativity) was frequently nonnuclear, whereas at term it was predominantly nuclear. Respective examples of both cases are indicated (arrowheads). Of note, we could not rule out the possibility that the cytoplasmic staining was nonspecific. If so, this would mean that the nuclear accumulation at term reflected new KDM6A synthesis; otherwise, it might in part reflect nuclear shuttling, as described previously for KDM6B (53). (G) Quantification of KDM6A distribution. Cells were scored as having KDM6A– or KDM6A+ nuclei based upon their nuclear-to-cytoplasmic staining ratio. n = 84–380 cells over at least 3 fields were scored per specimen. Error bars indicate SD. P values were determined by 2-tailed Mann-Whitney U test.
Discussion
We present evidence that uterine quiescence, the state of uterine noncontractility necessary to sustain pregnancy, is actively enforced by an H3K27me3-mediated gene-silencing program in DSCs. This program is induced in early gestation and influences decidual function in several ways. First, it inhibits type 1 immune responses, thus precluding the generation of associated inflammatory mediators such as TNF-α and PGE2, which promote myometrial contractility (16). This suppression anticipates alloimmune reactions to the placenta and has the added advantage of protecting the conceptus from rejection (7, 21). Second, the program suppresses myofibroblast formation and associated wound-healing responses, an unanticipated finding but one that makes eminent physiological sense given that implantation and the hemochorial mode of placentation seen in mice (and humans) entail severe disruptions to tissue integrity and hemostasis, including effacement of the uterine epithelium and trophoblast invasion into the decidual parenchyma and spiral arteries. Since myofibroblasts also produce inflammatory mediators and are themselves contractile (12–14), their generation within the decidua would also be at odds with the maintenance of uterine quiescence. Third, the program inhibits expression of Ptgfr and Oxtr. Intriguingly, signaling through FP, alone among all prostaglandin receptors, is profibrotic in the lung (22), while OXTR is upregulated by cancer-associated fibroblasts and its signaling can promote wound healing (23, 24). Thus, Ptgfr and Oxtr silencing could also be considered a feature of suppressed wound healing in the decidua.
We also provide evidence that relief from the gene-silencing program via H3K27 demethylation is required for labor entry, thus implicating H3K27me3 dynamics as an overarching regulator of the uterine state in mouse pregnancy (Figure 7). Indeed, given the upregulation of fibroblast activation markers with advancing gestation, parturition might be viewed as an exaggerated (albeit delayed) wound-healing response. This idea is consistent with findings that EZH2 inhibition promotes fibrosis (25, 26) and that PRC2 component downregulation, KDM6A/B upregulation, and H3K27me3 loss occur with wound healing (27). Of note, the consequences of H3K27 demethylation were evident as early as E15.5, 3 days prior to term; aside from the expansion in decidual macrophages that occurs during this period (28, 29) (and likely explained by our observation of Csf1 upregulation), changes this early might have been missed in other studies that focused on the terminal stages of pregnancy (29, 30). Interestingly, the lack of correlation between uterine H3K27me3 dynamics and uterine transcript levels for KDM6A/B and PRC2 components implicates posttranscriptional mechanisms as the ultimate determinants of H3K27 methylase and demethylase activities in DSCs. Also of interest, our observation that H3K27me3 did not appreciably accumulate on DSC>MSC target genes in the segments of undecidualized uteri between implantation sites suggests that P4, while possibly acting in a permissive fashion for H3K37me3 accumulation in early gestation DSCs, is not sufficient to induce H3K27me3 accumulation in undecidualized ESCs.
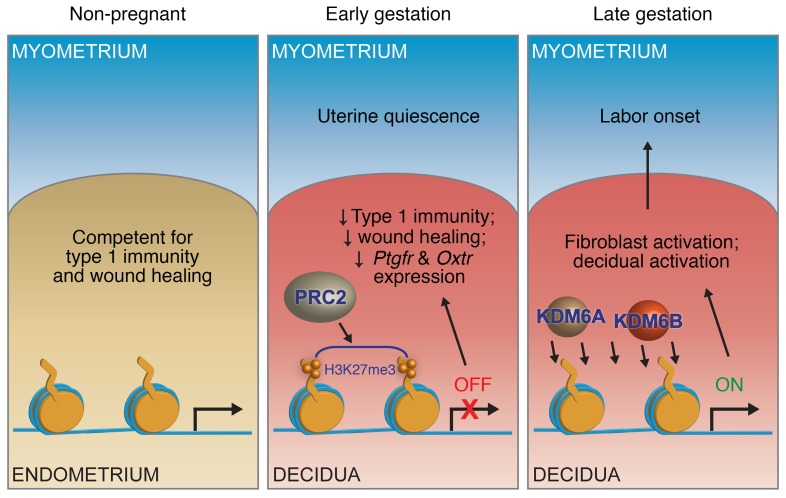
Figure 7. Proposed model for the role of H3K27me3 dynamics in pregnancy and parturition.
In early gestation, target gene accrual of H3K27me3 in DSCs inhibits the expression of genes that would otherwise promote uterine contractility and thus PTL; in late gestation, genome-wide H3K27 demethylation in DSCs, mediated by KDM6A and KDM6B, leads to target gene reexpression and decidual/fibroblast activation and thus in turn the generation of signals that promote myometrial contractility and labor entry.
Provocatively, we find that human DSCs downregulate α-SMA expression in early gestation, which suggests a potential commonality of gene silencing in both mouse and human DSCs as well as a shared need to avoid wound-healing responses in the decidua. Previous work also indicates α-SMA downregulation in baboon DSCs after an early phase of upregulation (31). Indeed, granulation tissue does not form within the human decidua, but the nonpregnant endometrium is competent for wound healing, as evidenced by Asherman syndrome, an endometrial scarring disorder that presents as infertility. Conversely, our findings that human DSCs at term manifest a myofibroblast-like phenotype and accumulate nuclear KDM6A suggest that epigenetic processes might also drive decidual activation and term parturition in humans rather than inflammation, as currently thought (2, 30, 32). The actual nature of H3K27me3 dynamics in human DSCs in vivo remains to be determined, however, with 2 genome-wide studies to date showing that H3K27me3 levels remain relatively constant or decline slightly when ESCs are decidualized in vitro over a 1- to 2-week period (33, 34). However, several considerations suggest that use of in vitro decidualization protocols may be inadequate for revealing H3K27me3 dynamics as they occur in vivo. First, it is unclear the extent to which in vitro–generated DSCs truly phenocopy their in vivo counterparts, even with regard to their transcriptional signature, let alone their epigenome. Indeed, DSCs are generated from ESCs in vitro through the use of medroxyprogesterone acetate (a nonmetabolizable progestin), frequently in combination with a cAMP analog and/or estrogen, whereas we found that the systemic hormonal environment of early pregnancy, which includes high P4 levels, is insufficient to induce H3K27me3 accrual in mouse ESCs in vivo, as discussed above. Second, it is possible that in vitro decidualization protocols generate “late gestation” rather than “early gestation” DSCs and so demonstrate H3K27me3 loss and consequent target gene upregulation rather than H3K27me3 accumulation and target gene silencing. Third, the genome-wide epigenetic changes that occur with adaptation to cell culture (e.g., see ref. 35) might preclude the generation of H3K27me3 marks in cultured DSCs.
That said, a direct parallel between mice and humans in terms of decidual H3K27me3 dynamics, if substantiated, suggests that parturition timing in uncomplicated pregnancy will be determined by developmental mechanisms that regulate the balance of H3K27 methylase and demethylase activities over the course of gestation, with events that occur even in early gestation having significant effects given that this is the stage when initial H3K27me3 levels in DSCs are established. This idea, in turn, suggests that preterm labor in the absence of infection might be the consequence of inappropriate exposure to epigenetic modifiers, either endogenous or environmental, acting potentially at any stage of gestation. Provocatively, idiopathic preterm labor has recently been associated with the choriodecidual upregulation of the human homologues of 2 H3K27me3 target genes, Tnfsf13b and Tslp, leading to B cell infiltration (36). Furthermore, the ability of GSK-J4 to prevent dinoprost-induced preterm labor without preventing luteolysis and consequently reduced plasma P4 suggests a link between decidual H3K27 demethylation and the functional P4 withdrawal also associated with idiopathic preterm labor. Further work will be necessary to clarify this issue as well as to determine whether decidual H3K27 demethylation creates a state of uterine P4 resistance at term.
These possibilities do not rule out a role for inflammation in term and preterm parturition. Indeed, decidual inflammation and leukocyte infiltration, viewed now as just one consequence of H3K27me3 demethylation in DSCs, could contribute to the terminal events of labor induction, including the initiation of myometrial contraction. This is consistent with the approximately 24-hour delays in parturition seen in various mouse strains with compromised inflammatory pathways (e.g., Il6–/– mice, ref. 30). Inflammatory mediators produced by the activated decidua might also further induce KDM6B (37), thus amplifying H3K27 demethylation in a feed-forward loop. In preterm labor associated with chorioamnionitis, the associated inflammatory response might itself be the stimulus that induces KDM6B expression and initiates the labor cascade. Although histone demethylases inhibitors per se might have contraindications for pregnancy use, these considerations together with the efficacy of GSK-J4 in mice inform what we believe to be a new paradigm for preterm labor management in humans.
Methods
Mouse husbandry and sources of uterine tissues.
Mice were housed in specific pathogen–free barrier facilities at NYU School of Medicine and UCSF. Pregnant mice were generated by mating B6CBAF1/J females (The Jackson Laboratory, 6 to 10 weeks of age) to C57BL/6 males (Taconic Farms). Noon of the day the copulation plug appeared was counted as E0.5. Artificially decidualized uteri were generated by mating B6CBAF1/J females with vasectomized ICR males (Taconic Farms) to induce pseudopregnancy and then injecting these females transcervically with 20 μl sesame seed oil on the equivalent of E3.5 as described previously (11). Nonpregnant uteri were obtained from B6CBAF1/J females given daily subcutaneous injections of 2 mg P4 (Sigma-Aldrich) dissolved in 100 μl sesame seed oil (Sigma-Aldrich) for 3 days prior to sacrifice.
Cell purification and culture.
USCs were isolated from total nonpregnant uteri (P4-treated mice); DSCs and MSCs were isolated from artificially decidualized uteri on the equivalent on E7.5 or from true implantation sites on E7.5 and E15.5. For DSCs and MSCs, uteri were first dissected to separate the decidua from overlying myometrium and then to separate the decidua from the placenta (E15.5 specimens). Embryos were discarded. Using our previously described protocol (7), the tissues and tissue layers were then enzymatically disaggregated with Liberase TM (Roche; 0.28 WU/ml), DNAase I (30 μg/ml), and trypsin (0.05%) in Ca++Mg++-containing HBSS for 60 minutes at 37°C with intermittent trituration. 5 mM EDTA was added for the final 15 minutes of the incubation. The ensuing cell suspensions were separated into stromal and nonstromal components through the use of LD magnetic bead columns (Miltenyi Biotec), according to the manufacturer’s protocol, and antibodies toward RBCs (Ter-119), leukocytes (CD45), epithelial cells (CD326) and endothelial cells (CD102). We used microbead-coupled goat anti-rat IgG antibodies (Miltenyi Biotec) as secondary reagents for the CD326 and CD102 antibodies, which were not directly bead coupled. Additional information on antibodies is in Supplemental Table 8. Stromal cells were routinely produced with approximately 95% purity, as assessed by flow cytometry for these same markers (7). The cells were cultured at a density of 0.5 × 106 cells/ml in SFM4CHO media supplemented with MEM nonessential amino acids, HEPES buffer, penicillin-streptomycin, l-glutamine, and sodium-pyruvate for a total of 24 hours prior to mRNA or chromatin isolation. When indicated, the cells were treated with a combination of 10 ng/ml mouse TNF-α (R & D Systems) and 1.000 U/ml mouse IFN-γ (Peprotech) for the last 6 hours of the culture period.
ChIP-Seq.
H3K27me3 ChIP was performed essentially as described previously (7). When applied to DSCs and MSCs (purified and cultured for 24 hours as described above; 0.5–1.0 × 106 cells/mouse), the cells were first fixed in 1% formaldehyde for 10 minutes at room temperature and then blocked with 0.125 M glycine for 5 minutes. When applied to whole-tissue extracts from pregnant mice on E7.5, implantation sites (5 to 7 per mouse) were first dissected from interimplantation sites (i.e., the segments of undecidualized uterus between implantation sites), and then the myometrium of each implantation site was dissected from the decidua. Embryos were discarded, and the tissues were then finely minced, fixed in 1% formaldehyde for 15 minutes at room temperature, blocked with 0.125 M glycine for 5 minutes, and then disaggregated with a dounce homogenizer on ice. In both cases, the fixed material was then washed twice in PBS, pelleted, and flash frozen for storage at –80°C. Thawed pellets were resuspended in cell-lysis buffer (5 mM pipes, pH 8.0, 85 mM KCl, 0.5% NP-40) containing fresh protease inhibitors (cOmplete Protease Inhibitor Cocktail, Roche), incubated for 10 minutes on ice, then centrifuged at 3,600 g at 4°C for 5 minutes. The nuclei were then lysed in nuclei-lysis buffer (50 mM Tris-HCL pH 8.0, 10 mM EDTA, 1% SDS, 1 mM 4-[2-aminoethyl] benzenesulfonyl fluoride hydrochloride [AEBSF], and cOmplete Roche protease inhibitors) for 10 minutes on ice. The chromatin was sonicated with a Bioruptor Plus to achieve an average DNA fragment length of 500 bp, as visualized by 1.5% agarose gel electrophoresis. Chromatin (10 μg) was precleared with 30 μl of blocked protein A/G beads (a 50:50 slurry of protein A– and G-sepharose [GE Healthcare] preincubated with 1 mg/ml BSA in 10 mM Tris pH 8.0, 1 mM EDTA). Chromatin was incubated overnight at 4°C with 2 μg H3K27me3 rabbit polyclonal antibodies (Millipore), then for 2 hours at 4°C with rotation in 35 μl of 50% blocked protein A/G beads. Beads and immune complexes were collected by centrifugation and successively washed 3 times with 1 ml low-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl pH 8.1, 150 mM NaCl), once with 1 ml high salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl pH 8.1, 500 mM NaCl), once with 1 ml lithium wash buffer (0.25 M LiCl, 1% NP-40, 1% sodium deoxycholate, 1 mM EDTA, 10 mM Tris-HCl pH 8.1), and twice with 1 ml TE8. The protein-DNA complexes were then eluted by incubating the beads in 150 μl elution buffer (1% SDS, 0.1 M NaHCO3) at 65°C for 15 minutes twice with change of elution buffer. Input DNA was prepared in parallel by adding elution buffer to 1% of input chromatin. The formaldehyde crosslinks were reversed by incubation overnight at 65°C. The day after, the solution was incubated for 1 hour at 37°C after the addition of 2 μl of 10 mg/ml RNAse A, then at 55°C for 2 hours after the addition of 3 μl of 20 mg/ml proteinase K, then extracted with phenol/chloroform. The DNA was then precipitated in ethanol/NaCl/glycogen and resuspended in 100 μl of 10 mM Tris pH 8.0.
ChIP-Seq libraries were generated by the Genome Technology Center at the NYU School of Medicine using end repair, A-tailing, adaptor ligation (Illumina TruSeq system), and PCR amplification. AMPure XP beads were used for DNA cleaning in each step of the process. Libraries were sequenced on a HiSeq 2000 sequencer (Illumina) using the 50-bp single-read method. Sequencing results were demultiplexed and converted to FASTQ format using Illumina bcl2fastq software. Reads were aligned to the mouse genome (build mm10/GRCm38) with Bowtie2 (38) using local alignment. Only confidently mapped reads (mapping quality > 20) were retained. Duplicate reads were discarded using the Picard toolkit (http://broadinstitute.github.io/picard/). MACS version 2.1.0 (39) was utilized to perform broad peak calling independently for each replicate with a q value cutoff of 0.1. Samples had 20,600,000 to 49,900,000 confidently mapped reads (19,700,000 to 42,400,000 after removing duplicates). MACS was also used to generate the fragment pileup and control λ scores in bedGraph files, which were then converted to bigWig format using bedGraphToBigWig (UCSC Browser binary utilities) in order to visualize the tracks. Snapshots were prepared using the Integrative Genomics Viewer (IGV; http://software.broadinstitute.org/software/igv/). R Bioconductor package DiffBind (https://bioconductor.org/packages/release/bioc/html/DiffBind.html) was used for identifying sites that were differentially enriched between sample groups. The differential analysis method was set to use the DESeq2 R package (40) for more conservative full library size normalization and to avoid the assumption that most of the sites are not differentially enriched.
Summated peak concentrations for individual chromosomes were calculated by raising the DiffBind-normalized log2(concentration) of every called peak by the second power and then summating these values over each chromosome. Heatmaps of enrichment around transcription start site (TSS) were generated using ngs.plot (41).
RNA-Seq.
RNA was isolated from DSCs, MSCs, and USCs using RNeasy Mini Plus reagents (QIAGEN) and QIAshredder columns (QIAGEN) according to the manufacturer’s protocol. The samples were then processed by the Genome Technology Center at the NYU School of Medicine. Poly(A)+ RNA was enriched using magnetic oligo(dT)-containing beads (Invitrogen). cDNA was prepared, and strand-specific libraries were constructed using the dUTP method. Libraries were sequenced on a HiSeq 2000 sequencer (Illumina) using the 50-bp paired-end method. Reads were aligned to the mouse genome (build mm10/GRCm38) using the splice-aware STAR aligner (42). PCR duplicates were removed using the Picard toolkit. HTSeq package (43) was utilized to generate counts for each gene based on how many aligned reads overlapped its exons. These counts were then normalized and used to test for differential expression using negative binomial generalized linear models implemented by the DESeq2 R package (40). Samples had 17,600,000 to 47,400,000 uniquely mapped reads. RNA-Seq was performed on 3 independent samples per group.
qRT-PCR.
Whole-tissue RNA was isolated using TRIzol (Thermo Fisher Scientific), and cDNA was synthesized from 1 μg RNA using an iScript cDNA Synthesis Kit (Bio-Rad). The PCR reaction was performed with Evagreen dye (Biotium) using a CFX Connect Real-Time PCR Detection System (Bio-Rad). qRT-PCR primers are listed in Supplemental Table 9.
Western blots.
Cells were lysed in 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% glycerol, 0.02% Igepal, and 2 mM EDTA, supplemented with complete Protease Inhibitor Cocktail (Roche), PMSF, α-glycerophosphate, NaF, and Na3VO4. The samples were spun at 3,000 g to remove debris, and 10 μg protein was loaded per well. Antibodies were diluted in PBS plus 1% BSA. Primary rabbit polyclonal antibodies were specific for H3K27me3 (Millipore), H3K9me3 (Abcam), and total histone H3 (Abcam). HRP-conjugated goat anti-rabbit antibodies were used as secondary reagents (Abcam). After each step, membranes were washed twice in 0.5% BSA/0.1% Tween/PBS and one time in PBS. HRP reactions were detected with an ECL kit (Bio-Rad). Images were acquired with a ChemiDoc Touch Imaging System (Bio-Rad). Densitometry was performed using ImageJ software and its gel analysis tool (NIH). Bands of interest were selected with the rectangle tool drawn as wide as possible without causing overlap with bands from adjacent lanes. For each lane, H3K27me3 or H3K9me3 levels were normalized to total histone H3 levels. To further normalize data across multiple independent gels, average total H3 density for the duplicate E7.5 lanes in a gel was set to 1. The same membranes were stripped in 1% SDS and 0.2 N NaOH and sequentially used for H3K27me3, H3K9me3, and total histone H3 detection. See complete unedited blots in the supplemental material.
H3K27me3 demethylase enzyme assays.
Nuclear extracts were prepared from the indicated tissues or tissue layers using a dounce homogenizer and a nuclear extraction kit according to the manufacturer’s instructions (Abcam, ab113474). We used 0.25–0.3 ml of the extraction buffer for each tissue layer from 4 implantation sites per mouse. Protein concentrations were determined by Bradford assay (Bio-Rad). After 10× dilution in water, the H3K27me3 demethylase assay was then performed on 0.5 μg nuclear extract using a commercial kit (Abcam, ab156910).
Tissue immunostaining.
Mouse tissues were fixed overnight at 4°C in 4% paraformaldehyde/PBS and embedded in paraffin. Sections were cut at 5 μm, baked 30 minutes at 65°C, then deparaffinized in xylene and ethanol using standard methods. The slides were incubated in methanol for 5 minutes, air-dried for 30 minutes, and then treated with 3% H2O2/0.1% sodium azide in PBS for 30 minutes. The sections were then subjected to antigen retrieval either using trypsin (1 mg/ml in PBS for 11 minutes at 37°C) or a Biocare NxGen decloaking chamber at 110°C for 30 seconds. They were then allowed to cool down for 10 minutes in the chamber, followed by 5 minutes at room temperature. The sections were then blocked in 1% BSA/3% donkey serum/0.004% Triton X-100 for 1 hour at room temperature, followed by application of primary antibodies, diluted in 1% BSA/0.4% Triton X-100/PBS. Antibody sources, dilutions, and antigen-retrieval methods are listed in Supplemental Table 8.
The immunostaining of mouse tissues relied upon biotin tyramide amplification for all antibodies except those against cytokeratin 8 (CK8) and α-SMA. The immunostaining of human tissues did not employ biotin tyramide amplification for any antibody except anti-KDM6A. After incubation with the primary antibody overnight at 4°C, samples with antibodies requiring amplification were next incubated with HRP-conjugated secondary antibodies (Jackson ImmunoResearch) diluted 1:200 in TNB buffer (PerkinElmer) for 30 minutes at room temperature, followed by biotin-tyramide amplification for 5 minutes at room temperature, then incubated with streptavidin–Alexa Fluor 594 (1:200; ThermoFisher Scientific) in 1% BSA/PBS for 30 minutes at room temperature. Afterward, they were mounted using DAPI–Fluoromount-G mounting media (Electron Microscopy Sciences). The slides were washed 3 times in PBS between all of these steps. Alexa Fluor 488– or Alexa Fluor 594–conjugated antibodies (1:200, Jackson Immunoresearch) were used as secondary reagents when amplification was not performed. Microscopy was performed using a Zeiss AxioImager M2 fitted with a motorized stage for automated image collection and an AxioCam 503 mono digital camera. Images at ×10 magnification collected with FITC, Texas red, and DAPI filter sets were tiled together to generate panoramic views using Zen software. Whenever staining intensity was the important parameter (EZH2, α-SMA, CXCL12, PDGFRB, KDM6A, CSPG4, F2RL1, FAP), the slides were stained at the same time and constant exposure times were used for the photomicroscopy. Adobe Photoshop was sometimes used to increase image brightness using the brightness/contrast function; in these cases, the alteration was applied evenly over the entire image and was applied identically to every image within the relevant experimental set. RBCs were visualized using long exposure times.
Lentiviral gene transfer.
The pWPI lentiviral vector (Addgene, no. 12254) containing a Csf1 cDNA insert was used to generate VSVG-pseudotyped lentiviral particles using standard methods. 75 × 107 Csf1-expressing or empty vector control virus mixed with 30 × 107 lentiviral particles produced from the FG12 EGFP reporter construct (Addgene, no. 14884), was injected into artificially decidualized uteri of pseudopregnant mice at multiple locations on E5.5 via a transmural approach, using our previously described methods (7). Each mouse received a total of 40 μl virus.
Determination of macrophage tissue densities.
After staining serial uterine sections for GFP and F4/80, each area of GFP positivity, along with the corresponding area from the anti-F4/80–stained section, was photomicrographed at 10× magnification. The images were then overlaid in Adobe Photoshop, the perimeter of the GFP+ area was outlined, and the number of F4/80+ cells within this perimeter was enumerated. At least 0.06 mm2 of GFP+ decidua were analyzed per mouse. F4/80+ cell densities in uninfected decidual areas of control virus–injected mice were calculated from at least 0.13 mm2 per mouse.
Wounding experiments.
Specimens for the analysis of the wounded artificial decidua came from the same mice that received control EGFP reporter lentiviruses via transmural injection (see above). Wounding of nondecidualized uteri employed the same surgical approach, but the inserted needle was then used to make a longitudinal scratch along the inner surface of the endometrium. Only one uterine horn was scratched; the other served as the nonwounded control. The mice received daily subcutaneous injections of 2 mg P4 dissolved in 100 μl sesame seed oil starting 6 hours after wounding, to simulate the hormonal environment of pregnancy and pseudopregnancy, and were sacrificed 2 days after wounding.
Parturition timing experiments.
Parturition timing and preterm labor experiments were performed at NYU School of Medicine. In this facility, parturition naturally occurred between midnight and 8 am on E18.5. For noninduced parturition experiments, pregnant B6CBAF/1 mice were given daily intraperitoneal injections of 80 μl vehicle (25% DMSO in PBS) or 25 mg/kg GSK-J4 (Cayman Chemical, 1797983-09-5) in vehicle starting on E13.5. Some mice were additionally given daily subcutaneous injections of 2 mg P4 in 100 μl sesame seed oil starting on E16.5. Mice were checked twice daily (6 pm and 9 am, counted as EX.5 or EX.0 in Figure 6E) for evidence of delivery, uterine bleeding, or maternal distress, which, if it occurred, caused us to sacrifice the mouse. For preterm labor experiments, mice were given GSK-J4 as above starting on E13.5 and then given a single intraperitoneal injection of 60 μg synthetic PGF2α (dinoprost, Cayman Chemical, 551-11-1) dissolved in 80 μl 25% DMSO/PBS on E16.5 with or without concomitant P4 injection, and sacrificed on E17.5.
P4 measurements.
Plasma P4 concentrations were determined using a commercial EIA kit (Cayman Chemical). Mice were anesthetized and blood was collected by cardiac puncture into heparinized tubes. Plasma was stored at –80°C until use. Duplicate measurements were performed for each sample and then averaged.
Human specimens.
Formalin-fixed, paraffin-embedded human specimens were obtained via clinical database searches. Placental/decidual specimens from 6- and 13-week elective terminations of pregnancy, performed via uterine aspiration without mifepristone administration, were identified within the Pathology Department database of Zuckerberg San Francisco General Hospital (San Francisco, California, USA). Term placental specimens (37 to 39 weeks of gestation), characterized by clinical indication for nonlaboring, elective cesarean section delivery, were identified within the Pathology Department database of UCSF. H&E-stained slides were screened by a pathologist (GR) to confirm an absence of maternal and fetal pathology (including chorioamnionitis, meconium, villitis, chronic deciduitis or other pathological inflammation, vascular pathology, maturation or perfusion defects) and to select blocks containing adequate amounts of decidual tissue.
Data availability.
ChIP-Seq and RNA-Seq data sets were deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE105456).
Statistics.
Comparisons of H3K27me3 peak concentrations between DSC>MSC and MSC>DSC subsets (Figure 1D and Supplemental Figure 1C) were analyzed by unpaired t test, as were qRT-PCR and lentiviral experiments. The qRT-PCR analysis employed raw ΔCt values. All t tests were 2 tailed. All analyses that employed 1-way ANOVA were followed by Tukey’s multiple comparison test unless otherwise stated. For parametric analyses of data sets with small sample sizes, normality was assumed after visual inspection of the data ruled out a high degree of skewing and in consideration of the fact that the experiments were performed identically on genetically identical mice. In these cases, parametric tests were also considered appropriate because they provide an acceptable level of about 5% false positives for nonnormal data unless the data are highly skewed (44). We used the hypergeometric test available with the online Keisan calculator (http://keisan.casio.com/exec/system/1180573201) to determine whether a gene subset was significantly over- or underrepresented within a larger gene set; for Figure 3, A–C, this included Bonferroni’s correction for the 9 different gene-expression bins, giving a threshold P value for significance of 5.6 × 10–3, but otherwise P < 0.05 was considered significant. Parturition timing experiments were analyzed by the log-rank (Mantel-Cox) test. Additional tests are described in the main text and figure legends. All analyses aside from ChIP-Seq, RNA-Seq, and hypergeometric calculations were performed using GraphPad Prism software, and P < 0.05 was considered significant. For ChIP-Seq and RNA-Seq analyses, FDR < 0.05 and Padj < 0.05, respectively, were considered significant. There was no blinding, and no data were excluded from analysis for nontechnical reasons.
Study approval.
All animal experiments were approved by the Institutional Animal Care and Use Committees of NYU School of Medicine and UCSF. Formalin-fixed, paraffin-embedded human specimens were obtained through a protocol (CHR#12-09332) approved by the UCSF IRB that allows for archived specimens to be used for research purposes after deidentification.
Author contributions
PN, JS, and GR conceived of and performed experiments, analyzed data, and wrote the manuscript. ET, IO, PM, and CC performed experiments. ID and AT performed the bioinformatics analysis. AE conceived of the project, analyzed data, wrote the manuscript, and provided supervision.
Supplementary Material
Acknowledgments
We thank Nataliya Korets, Chin-Siean Tay, and Devin Columbus for technical assistance and Susan Fisher and Linda Guidice for discussions. The Histopathology and Genome Technology cores of NYU School of Medicine were supported by NIH grant P30CA016087. The Applied Bioinformatics Core and the High Performance Computing Facility of NYU School of Medicine provided bioinformatics support. This work was supported by grants from the NIH (R01AI106745) and the March of Dimes (#6-FY13-80) to AE.
Version 1. 11/27/2017
Electronic publication
Version 2. 11/27/2017
Corrected supplemental data files.
Version 3. 11/28/2017
Added table of contents to supplemental data file.
Version 4. 01/02/2018
Print issue publication
Footnotes
AE’s present address is: Department of Laboratory Medicine, UCSF, San Francisco, California, USA.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2018;128(1):233–247.https://doi.org/10.1172/JCI95937.
Contributor Information
Patrice Nancy, Email: patricenancy@gmail.com.
Johan Siewiera, Email: JohanPierreRobert.Siewiera@ucsf.edu.
Gabrielle Rizzuto, Email: Gabrielle.Rizzuto@ucsf.edu.
Elisa Tagliani, Email: elisa.tagliani5@gmail.com.
Ivan Osokine, Email: ivan.osokine@ucsf.edu.
Priyanka Manandhar, Email: Priyanka.Manandhar@ucsf.edu.
Igor Dolgalev, Email: igor.dolgalev@nyumc.org.
Caterina Clementi, Email: c.clementi@celmatix.com.
Aristotelis Tsirigos, Email: aristotelis.tsirigos@nyumc.org.
Adrian Erlebacher, Email: adrian.erlebacher@ucsf.edu.
References
- 1.Cha J, Sun X, Dey SK. Mechanisms of implantation: strategies for successful pregnancy. Nat Med. 2012;18(12):1754–1767. doi: 10.1038/nm.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shynlova O, Lee YH, Srikhajon K, Lye SJ. Physiologic uterine inflammation and labor onset: integration of endocrine and mechanical signals. Reprod Sci. 2013;20(2):154–167. doi: 10.1177/1933719112446084. [DOI] [PubMed] [Google Scholar]
- 3.Norwitz ER, et al. Molecular regulation of parturition: the role of the decidual clock. Cold Spring Harb Perspect Med. 2015;5(11):a023143. doi: 10.1101/cshperspect.a023143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Romero R, Dey SK, Fisher SJ. Preterm labor: one syndrome, many causes. Science. 2014;345(6198):760–765. doi: 10.1126/science.1251816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muglia LJ, Katz M. The enigma of spontaneous preterm birth. N Engl J Med. 2010;362(6):529–535. doi: 10.1056/NEJMra0904308. [DOI] [PubMed] [Google Scholar]
- 6.Hirota Y, Cha J, Dey SK. Revisiting reproduction: Prematurity and the puzzle of progesterone resistance. Nat Med. 2010;16(5):529–531. doi: 10.1038/nm0510-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nancy P, Tagliani E, Tay CS, Asp P, Levy DE, Erlebacher A. Chemokine gene silencing in decidual stromal cells limits T cell access to the maternal-fetal interface. Science. 2012;336(6086):1317–1321. doi: 10.1126/science.1220030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Griffith JW, Sokol CL, Luster AD. Chemokines and chemokine receptors: positioning cells for host defense and immunity. Annu Rev Immunol. 2014;32:659–702. doi: 10.1146/annurev-immunol-032713-120145. [DOI] [PubMed] [Google Scholar]
- 9.Tagliani E, Shi C, Nancy P, Tay CS, Pamer EG, Erlebacher A. Coordinate regulation of tissue macrophage and dendritic cell population dynamics by CSF-1. J Exp Med. 2011;208(9):1901–1916. doi: 10.1084/jem.20110866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Redline RW, Lu CY. Specific defects in the anti-listerial immune response in discrete regions of the murine uterus and placenta account for susceptibility to infection. J Immunol. 1988;140(11):3947–3955. [PubMed] [Google Scholar]
- 11.Collins MK, Tay CS, Erlebacher A. Dendritic cell entrapment within the pregnant uterus inhibits immune surveillance of the maternal/fetal interface in mice. J Clin Invest. 2009;119(7):2062–2073. doi: 10.1172/JCI38714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Augsten M. Cancer-associated fibroblasts as another polarized cell type of the tumor microenvironment. Front Oncol. 2014;4:62. doi: 10.3389/fonc.2014.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hinz B, et al. Recent developments in myofibroblast biology: paradigms for connective tissue remodeling. Am J Pathol. 2012;180(4):1340–1355. doi: 10.1016/j.ajpath.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stempien-Otero A, Kim DH, Davis J. Molecular networks underlying myofibroblast fate and fibrosis. J Mol Cell Cardiol. 2016;97:153–161. doi: 10.1016/j.yjmcc.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sugimoto Y, et al. Failure of parturition in mice lacking the prostaglandin F receptor. Science. 1997;277(5326):681–683. doi: 10.1126/science.277.5326.681. [DOI] [PubMed] [Google Scholar]
- 16.Christiaens I, Zaragoza DB, Guilbert L, Robertson SA, Mitchell BF, Olson DM. Inflammatory processes in preterm and term parturition. J Reprod Immunol. 2008;79(1):50–57. doi: 10.1016/j.jri.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 17.Arcipowski KM, Martinez CA, Ntziachristos P. Histone demethylases in physiology and cancer: a tale of two enzymes, JMJD3 and UTX. Curr Opin Genet Dev. 2016;36:59–67. doi: 10.1016/j.gde.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kokubu K, Hondo E, Sakaguchi N, Sagara E, Kiso Y. Differentiation and elimination of uterine natural killer cells in delayed implantation and parturition mice. J Reprod Dev. 2005;51(6):773–776. doi: 10.1262/jrd.17033. [DOI] [PubMed] [Google Scholar]
- 19.Elovitz MA, Mrinalini C. Animal models of preterm birth. Trends Endocrinol Metab. 2004;15(10):479–487. doi: 10.1016/j.tem.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 20.Franquemont DW, Frierson HF, Jr , and Mills SE. An immunohistochemical study of normal endometrial stroma and endometrial stromal neoplasms. Evidence for smooth muscle differentiation. Am J Surg Pathol. 1991;15(9):861–870. doi: 10.1097/00000478-199109000-00006. [DOI] [PubMed] [Google Scholar]
- 21.Arck PC, Hecher K. Fetomaternal immune cross-talk and its consequences for maternal and offspring’s health. Nat Med. 2013;19(5):548–556. doi: 10.1038/nm.3160. [DOI] [PubMed] [Google Scholar]
- 22.Oga T, et al. Prostaglandin F(2alpha) receptor signaling facilitates bleomycin-induced pulmonary fibrosis independently of transforming growth factor-beta. Nat Med. 2009;15(12):1426–1430. doi: 10.1038/nm.2066. [DOI] [PubMed] [Google Scholar]
- 23.Navab R, et al. Prognostic gene-expression signature of carcinoma-associated fibroblasts in non-small cell lung cancer. Proc Natl Acad Sci U S A. 2011;108(17):7160–7165. doi: 10.1073/pnas.1014506108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poutahidis T, et al. Microbial symbionts accelerate wound healing via the neuropeptide hormone oxytocin. PLoS ONE. 2013;8(10):e78898. doi: 10.1371/journal.pone.0078898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou X, et al. Enhancer of zeste homolog 2 inhibition attenuates renal fibrosis by maintaining Smad7 and phosphatase and tensin homolog expression. J Am Soc Nephrol. 2016;27(7):2092–2108. doi: 10.1681/ASN.2015040457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krämer M, et al. Inhibition of H3K27 histone trimethylation activates fibroblasts and induces fibrosis. Ann Rheum Dis. 2013;72(4):614–620. doi: 10.1136/annrheumdis-2012-201615. [DOI] [PubMed] [Google Scholar]
- 27.Shaw T, Martin P. Epigenetic reprogramming during wound healing: loss of polycomb-mediated silencing may enable upregulation of repair genes. EMBO Rep. 2009;10(8):881–886. doi: 10.1038/embor.2009.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamilton S, et al. Macrophages infiltrate the human and rat decidua during term and preterm labor: evidence that decidual inflammation precedes labor. Biol Reprod. 2012;86(2):39. doi: 10.1095/biolreprod.111.095505. [DOI] [PubMed] [Google Scholar]
- 29.Shynlova O, Nedd-Roderique T, Li Y, Dorogin A, Nguyen T, Lye SJ. Infiltration of myeloid cells into decidua is a critical early event in the labour cascade and post-partum uterine remodelling. J Cell Mol Med. 2013;17(2):311–324. doi: 10.1111/jcmm.12012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robertson SA, et al. Interleukin-6 is an essential determinant of on-time parturition in the mouse. Endocrinology. 2010;151(8):3996–4006. doi: 10.1210/en.2010-0063. [DOI] [PubMed] [Google Scholar]
- 31.Christensen S, et al. Smooth muscle myosin II and alpha smooth muscle actin expression in the baboon (Papio anubis) uterus is associated with glandular secretory activity and stromal cell transformation. Biol Reprod. 1995;53(3):598–608. doi: 10.1095/biolreprod53.3.598. [DOI] [PubMed] [Google Scholar]
- 32.Stephen GL, et al. Transcriptomic profiling of human choriodecidua during term labor: inflammation as a key driver of labor. Am J Reprod Immunol. 2015;73(1):36–55. doi: 10.1111/aji.12328. [DOI] [PubMed] [Google Scholar]
- 33.Grimaldi G, Christian M, Steel JH, Henriet P, Poutanen M, Brosens JJ. Down-regulation of the histone methyltransferase EZH2 contributes to the epigenetic programming of decidualizing human endometrial stromal cells. Mol Endocrinol. 2011;25(11):1892–1903. doi: 10.1210/me.2011-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tamura I, et al. Genome-wide analysis of histone modifications in human endometrial stromal cells. Mol Endocrinol. 2014;28(10):1656–1669. doi: 10.1210/me.2014-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nestor CE, et al. Rapid reprogramming of epigenetic and transcriptional profiles in mammalian culture systems. Genome Biol. 2015;16:11. doi: 10.1186/s13059-014-0576-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang B, et al. Interleukin-33-induced expression of PIBF1 by decidual B cells protects against preterm labor. Nat Med. 2017;23(1):128–135. doi: 10.1038/nm.4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130(6):1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 38.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shen L, Shao N, Liu X, Nestler E. ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics. 2014;15:284. doi: 10.1186/1471-2164-15-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. McDonald JH. Handbook of Biological Statistics. Baltimore, Maryland: Sparky House Publishing; 2014. [Google Scholar]
- 45.Su Y, Nishimoto T, Feghali-Bostwick C. IGFBP-5 promotes fibrosis independently of its translocation to the nucleus and its interaction with nucleolin and IGF. PLoS One. 2015;10(6):e0130546. doi: 10.1371/journal.pone.0130546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Surendran K, Simon TC. CNP gene expression is activated by Wnt signaling and correlates with Wnt4 expression during renal injury. Am J Physiol Renal Physiol. 2003;284(4):F653–F662. doi: 10.1152/ajprenal.00343.2002. [DOI] [PubMed] [Google Scholar]
- 47.Allaoui R, et al. Cancer-associated fibroblast-secreted CXCL16 attracts monocytes to promote stroma activation in triple-negative breast cancers. Nat Commun. 2016;7:13050. doi: 10.1038/ncomms13050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chung H, Ramachandran R, Hollenberg MD, Muruve DA. Proteinase-activated receptor-2 transactivation of epidermal growth factor receptor and transforming growth factor-β receptor signaling pathways contributes to renal fibrosis. J Biol Chem. 2013;288(52):37319–37331. doi: 10.1074/jbc.M113.492793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saliba Y, et al. Evidence of a role for fibroblast transient receptor potential canonical 3 Ca2+ channel in renal fibrosis. J Am Soc Nephrol. 2015;26(8):1855–1876. doi: 10.1681/ASN.2014010065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Millar NL, O’Donnell C, McInnes IB, Brint E. Wounds that heal and wounds that don’t - The role of the IL-33/ST2 pathway in tissue repair and tumorigenesis. Semin Cell Dev Biol. 2017;61:41–50. doi: 10.1016/j.semcdb.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 51.De Langhe E, Aznar-Lopez C, De Vooght V, Vanoirbeek JA, Luyten FP, Lories RJ. Secreted frizzled related proteins inhibit fibrosis in vitro but appear redundant in vivo. Fibrogenesis Tissue Repair. 2014;7:14. doi: 10.1186/1755-1536-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hanna J, et al. CXCL12 expression by invasive trophoblasts induces the specific migration of CD16- human natural killer cells. Blood. 2003;102(5):1569–1577. doi: 10.1182/blood-2003-02-0517. [DOI] [PubMed] [Google Scholar]
- 53.Kamikawa YF, Donohoe ME. The localization of histone H3K27me3 demethylase Jmjd3 is dynamically regulated. Epigenetics. 2014;9(6):834–841. doi: 10.4161/epi.28524. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ChIP-Seq and RNA-Seq data sets were deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE105456).