Abstract
Background
There is conflicting data on the role of macrophages in colorectal cancer (CRC); some studies have shown that macrophages can exert an anti-tumor effect whereas others show that macrophages are tumor promoting. We sought to determine the role of conditioned medium (CM) from macrophages, in particular classically activated macrophages, on the development of the CSC phenotype in CRC cells, which is believed to mediate tumor growth and chemoresistance.
Methods
Murine (CT26) and human (HCP-1) CRC cell lines were treated with CM from lipopolysaccharide (LPS)-activated murine macrophages. The CSC population was assessed using the sphere-forming assay and aldehyde dehydrogenase assay. Chemoresistance studies were performed using the MTT assay. CSC transcription factors and SHH protein were analyzed by Western blotting.
Results
The results showed that LPS-activated macrophage CM induced the CSC phenotype in CRC cells. Further studies showed that the CSC phenotype was mediated by the sonic hedgehog (SHH)-Gli signaling pathway, which is known to drive self-renewal; these effects were blocked by depletion of SHH in macrophage CM. In addition, LPS-activated macrophage CM enhanced chemoresistance.
Conclusions
LPS-activated macrophages play an active role in promoting the CSC phenotype through activation of the SHH-Gli signaling pathway in CRC cells.
Introduction
Colorectal cancer (CRC) is the second leading cause of cancer-related death in the United States [1]. Several clinical trials have led to the approval of new drugs for patients with metastatic CRC (mCRC) within the past few years; however, the prognosis remains poor, and most patients will develop resistance to therapy within one year [1–3]. Thus, finding method to combat resistance to therapy is critical if we are to improve the survival of patients with mCRC.
Cancer stem cells (CSCs), a subpopulation of cancer cells that are thought to drive tumor growth and metastasis, have been identified in multiple types of malignancies, including CRC [4–13]. CSCs are thought to not only initiate and sustain tumor growth but also mediate chemoresistance [14–20], mostly through key signaling pathways such as Wnt, Notch, and Hedgehog [21]. Notably, results from our laboratory and others suggest that CSCs exist in a state of flux and that the CSC phenotype can be enhanced by micro-environmental influences [22–26]. Currently most therapies targeting the bulk of the tumor cells ultimately fail as they do not successfully eliminate CSCs. Therefore, understanding the CSC-microenvironment relationship and the development of CSC- or microenvironment-targeting strategies that could eliminate or deplete the CSC population are critical for improving the clinical outcomes of patients with mCRC.
The tumor microenvironment is complex and made up of various cell types, including tumor associated macrophages (TAMs) (reviewed in [27]). Macrophages have been previously shown to interact with stem cells [28], and a growing body of evidence suggests that TAMs are critical for the self-renewal and maintenance of CSCs in established tumors [29–31]. Recent studies have sought to characterize the relationship and cross-talk between TAMs and CSCs (reviewed in [30]) and have demonstrated that TAMs are able to increase the number of CSCs and cell phenotypes in hepatocellular carcinoma and pancreatic ductal carcinoma [32–34]. Also, it was shown that TAMs can increase both tumorigenicity and drug resistance in CRC both in vitro and in vivo [35]. These effects were due to coordinated activation of Stat3 and Sonic Hedgehog (SHH) signaling in CRC stem cells. However, despite these exciting results, there is still a tremendous amount to learn about the interactions between TAMs and CSCs, particularly in CRC.
Typically, macrophages are divided into two main classes, M1 (classically activated macrophages; antitumor activities) and M2 (alternatively activated-macrophages: protumor activities) [29, 36]. However, the role of different macrophages in the CRC microenvironment are not well defined and remains a point of controversy (reviewed in [37–39]). In vitro, exposure to lipopolysaccharide (LPS) polarizes macrophages toward an M1 phenotype, whereas exposure to IL-4 or IL-13 polarizes them towards an M2 phenotype. In this study, we examined the effects of classically LPS-activated macrophage CM on the acquisition of the CSC phenotype in CRC cells. Our studies demonstrated that the CM of LPS-activated macrophages is able to increase the CSC phenotype and promote chemoresistance of CRC cells. We also showed that the CSC phenotype was enhanced by the secretion of sonic hedgehog (SHH) by LPS-activated macrophages.
Materials and methods
Cell lines
Freshly isolated HCP-1 CRC cells were established in our laboratory, as previously described [40]. The murine cell lines CT26 and RAW264.7 (hereafter RAW) and the human monocyte cell line U937 were purchased from American Type Culture Collection (Manassas, VA, USA). CT26 and RAW cells were maintained in culture using standard protocols in minimal essential medium, supplemented with 10% fetal bovine serum (FBS) at 37°C in 5% CO2. U937 cells were maintained in RPMI 1640 medium supplemented with 10% FBS, 2 mmol/L L-glutamine, and 0.05 mM 2-mercaptoethanol. Cells were confirmed to be free of mycoplasma using the MycoAlert mycoplasma detection kit (Lonza Group, Allendale, NJ). The results of all in vitro studies were reproduced in at least three independent experiments.
Macrophage differentiation
Human blood was obtained from healthy (anonymous) donors at the Gulf Coast Regional Blood Center, Houston TX, and was purchased the Blood Center with an IRB exemption. The monocytes were obtained from buffy coat by gradient centrifugation using Ficoll-Paque (GE Healthcare Life Sciences). Non-adherent cells were removed and purified monocytes were incubated for 7 days in RPMI 1640 supplemented with 10% FBS and 50 ng/ml M-CSF to obtain macrophages (hereafter Human Primary Macrophages). Cells were washed with PBS twice and incubated overnight with 10% FBS-MEM supplemented with 1 μg/ml of LPS (Sigma, St. Louis, MO, USA). Cells were then washed with PBS twice and cultured with MEM-1% FBS for 48 h. The conditioned medium was harvested and filtered through a 0.22-μm filter to remove cell debris before being added to the CRC cell cultures.
Conditioned medium preparation
CT26, HCP-1, and RAW cells were cultured under MEM-1% FBS conditions for 48 h. The media were harvested and filtered through a 0.22-μm filter to remove cell debris and serve as a control. Murine RAW macrophages and human U937 monocytes were activated using 1 μg/ml of LPS solution and incubation overnight. Cells were then washed with PBS twice and cultured with MEM-1% FBS for 48 h. The media were harvested and filtered through a 0.22-μm filter to remove cell debris before being added to the CRC cell cultures.
MTT assay
Pretreated CRC cells with CM for 48 h, cells were then trypsinized and seeded 3,000 cells/well with CM with or without 5FU or SN38 into the 96 well plates and the cells were incubated for 72 h. At the end of the incubation, 3- [4, 5-dimethyl-thiazol-2-yl] 2, 5 diphenyltetrazolium bromide (MTT; Sigma) was added to a final concentration of 0.5 mg/ml, and the cells were incubated for another 2 h. After the medium and MTT were removed, dimethyl sulfoxide was added for 1 min, and absorption was read at 570 nm.
Aldefluor assay
The Aldefluor kit from Stemcell Technologies (Vancouver, CA) was used to identify cells that exhibited high ALDH enzymatic activity, according to the manufacturer’s instructions. In brief, cells were trypsinized and suspended in Aldefluor assay buffer containing ALDH substrate (BAAA, 1 μmol/L) and incubated at 37°C for 30 minutes. As a negative control, an aliquot from each sample was treated with 50 mmol/L diethyl-aminobenzaldehyde, a specific ALDH inhibitor, and followed up by flow cytometric analysis using FlowJo software (Tree Star, Inc., Ashland, OR).
Sphere-forming assay
CT26 and HCP-1 cells were plated in 96-well, ultra-low-attachment plates (BD Biosciences, San Jose, CA) at a density of 50 or 100 viable cells per well, respectively. Standard sphere-forming medium (serum-free DMEM/F-12 supplemented with 1× B27 serum substitute, 20 ng/ml human recombinant epidermal growth factor, and 20 ng/ml basic fibroblast growth factor [all from Invitrogen, Carlsbad, CA]) was mixed at a 1:3 ratio with macrophage CM or control CM and added to the CRC cells. Plates were incubated at 37°C and 5% CO2 and cultured for 7–14 days. Spheres larger than 50 μm in diameter were counted. For siRNA knockdown, cells were transfected with siRNAs, recovered overnight in normal growth medium, and then the cells were single suspended and seeded 100 cells /well as described above. For assays with the smoothened (SMO) inhibitor, LDE225 (Selleckchem Houston, TX), cells were pretreated with the inhibitor at indicated doses for 4 hours, and then the cells were single suspended and seeded at 100 cells / well as described above with inhibitor.
Promoter reporter assay
HCP-1 cells were infected with lentiviruses containing a Hes-1 AB Notch/CSL luciferase reporter [41], a pWPT-Gli-GFP reporter, or a pWPT-TCF-GFP reporter (kind gifts from Dr. Renuka P. Limgala, NIH [42]). The reporter-containing CRC cells were then incubated with LPS-activated macrophage CM. After 24 h of culture, promoter activities were determined by FACS analysis.
Western blot analysis
Cell lysates or concentrated CM (concentrated by P3 Ultra-4 centrifugal filter units [Millipore]) were run on SDS-PAGE, following a standard protocol. The following primary antibodies were used: Gli, OCT/4, β-Catenin (Cell Signaling, Danvers, MA), Hes-1 (Abcam, Cambridge, MA), rabbit polyclonal antibody SHH, Nanog, and β-actin (Santa Cruz, Dallas, TX). Signals were detected by chemoluminescence (Fisher Scientific, Pittsburgh, PA).
Reverse transcription polymerase chain reaction
RNA from RAW or RAW+LPS cells was isolated by TRIzol extraction (Invitrogen) and purified using the RNeasy kit (Qiagen, Valencia, CA). First-strand cDNA was synthesized with SuperScript III reverse transcriptase (Invitrogen). PCR amplification for M1 or M2 macrophage markers was performed under the following conditions: 95°C for 5 min, 25 cycles of 30 s denaturation at 95°C, 30 s annealing at 60°C, and 1 minute of extension at 72°C. Products were analyzed by electrophoresis of 20 μL of each PCR reaction mixture in a 1.5% agarose gel, and bands were visualized by ethidium bromide staining. The primers used are presented in S1 Table.
siRNA knockdown
A mixture of two different small interfering RNAs (siRNAs) targeting SHH (designed by multiple siRNA design platforms) was used to deplete SHH in RAW cells. The sequences were #1 sense 5’ CAUCCACUGUUCUGUGAAA, antisense 5’ UUUCACAGAACAGUGGAUG; and #2 sense 5’ GGGUCUACUAUGAAUCCAA, antisense 5’ UUGGAUUCAUAGUAGACCC. A validated non-specific siRNA (control siRNA) was obtained from Sigma. Lipofectamine 2,000 siRNA transfection reagent was used according to the manufacturer’s instructions (Invitrogen). The siRNA knockdown was performed in Opti-MEM (Gibco). 24 h after transfection, cells were washed with PBS, cultured in MEM-10% FBS supplemented with LPS, and incubated overnight. Cells were then washed with PBS twice and cultured with MEM-1% FBS. 48 h later, supernatant media were collected and assessed for SHH depletion via Western blot analysis.
Statistical analyses
Statistical analyses were performed using Student’s t-test (Microsoft Excel 2007, Redmond, WA). All statistical tests were 2-sided, and P values ≤0.05 were considered statistically significant.
Result
Murine macrophage CM promotes the CRC stem cell phenotype in murine and human CRC cell lines
The RAW macrophages were first polarized by LPS into classically activated macrophages. The resulting population was characterized by RT-PCR through the detection of M1 and M2 polarization markers, as shown in S1 Table. Our PCR results showed that LPS-activated macrophages, although classically described as M1 macrophages, are actually a mixture of both M1-like and M2-like macrophage phenotypes (S1A Fig).
To understand the potential role of macrophage-secreted factors in promoting the CSC phenotype of CRC cells, we incubated CT26 and HCP-1 cells with CM obtained from LPS-activated RAW macrophages; in parallel, we performed stimulations with non-activated RAW and CRC cells, using their own CM as the controls. CRC cells were analyzed using two common techniques for detecting the CSC phenotype: sphere-forming and ALDH activity assays. Fig 1A and 1B show that LPS-activated macrophage CM increased the sphere-forming capability of both murine CT26 and human HCP-1 CRC cell lines by more than two-fold compared to the control CM. In addition, LPS-activated macrophage CM treatment enriched the Aldefluor-positive cell population of both CT26 and HCP-1 cells by ~four- and ~eight-fold, respectively, compared to controls (Fig 1C and 1D). As a secondary readout for stem-ness, we also assessed alterations in CD133 positive CRC cells in response to treatment with LPS-activated macrophage CM. Exposure to LPS-activated macrophage CM led to a ~3-fold increase in CD133 positive CRC cells as compared to control or untreated macrophages (data not shown). Together, these data suggest that LPS-activated macrophage-derived soluble factors promote the CSC phenotype of CRC cells in a paracrine manner.
Fig 1. Murine macrophage Conditioned Media (CM) promotes the stem cell phenotype of CRC cells.
(A and B) CT26 and HCP-1 cells were cultured with LPS-activated macrophage CM or control CM, and a sphere-forming assay was performed (*P < 0.01). (C and D) Aldefluor-positive cell population was determined (*P < 0.001).
Murine macrophage CM enhanced the CSC phenotype through the SHH signaling pathway
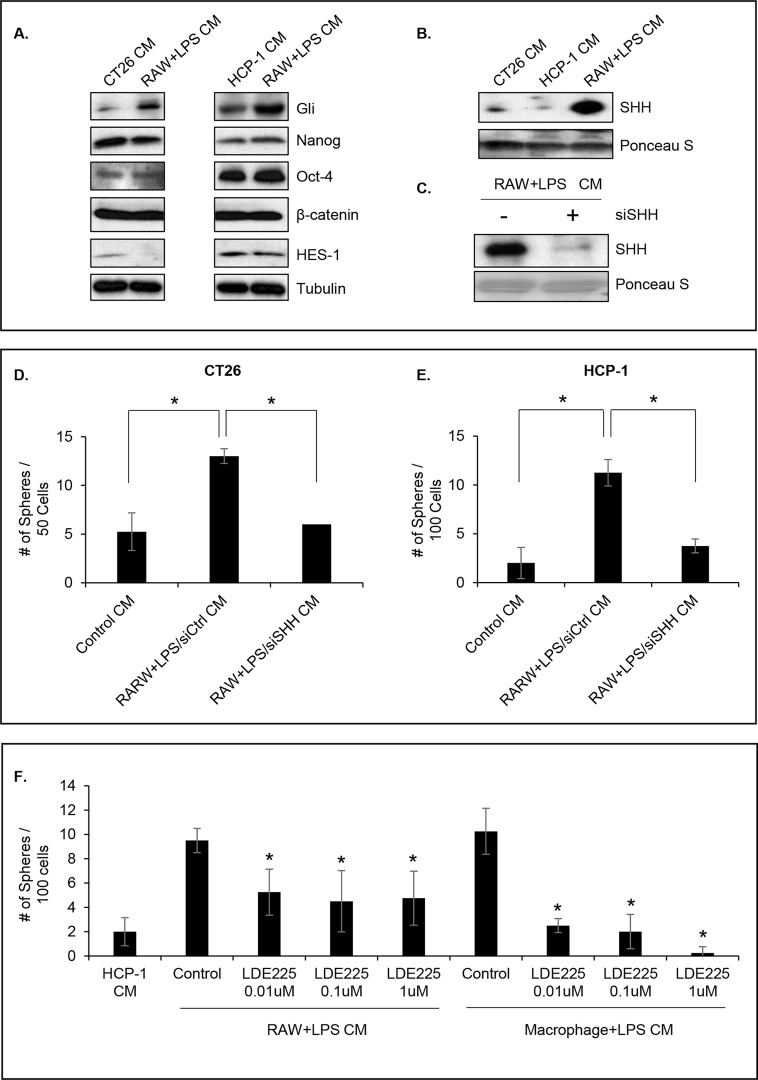
To understand the underlying mechanism of the activation of the CSC phenotype in CRC cells by LPS-activated macrophage CM, we determined the activity of canonical CSC pathways such as Notch [43], Wnt/β-Catenin [44], and SHH [45] in these cells. To this end, we infected HCP-1 cells with a lentivirus containing a Hes-1, TCF, or Gli promoter-driven reporter and treated them with LPS-activated macrophage CM or control CM for 48 h. LPS-activated macrophage CM treatment led to an increase in the activity of Gli promoter but not of Hes-1 or TCF promoter (S2 Fig). In addition, Western blot analysis of the canonical CSC transcription factors (Gli, HES-1, β-Catenin, Nanog, and OCT-4) confirmed that Gli protein levels were significantly increased following LPS-activated macrophage CM treatment of both CT26 and HCP-1 cells (Fig 2A). Moreover, since the Gli pathway is activated by SHH, we assessed the presence of SHH in CM by Western blot analysis. The data demonstrated that SHH was secreted at high levels by LPS-activated macrophages compared to CT26 and HCP-1 cells (Fig 2B) and suggest that LPS-activated macrophages secrete SHH that, in turn, activates the SHH intracellular pathway in CRC in a paracrine fashion. Of note, there is 92.4% shared identity between human and mouse SHH proteins [46]; therefore, one should not expect inter-species protein interactions to be an issue in the following experiments.
Fig 2. Murine macrophage CM enhanced the SHH signaling pathway.
(A) CSC transcription factor expression with 48 h of CM treatment in CT26 and HCP-1 cells. (B) Western blot analysis of SHH secretion in CRC cells and RAW cells. (C) Western blot analysis shows knockdown of SHH at 48 h after the indicated siRNA transfections. (D and E) Effects of knockdown SHH-mediated sphere formation in CT26 and HCP-1 cells (*P < 0.01). (F) HCP-1 cells were cultured with LPS-activated RAW CM or LPS-activated primary macrophage CM with or without a SMO inhibitor and the sphere-forming assay was performed (*P < 0.01).
To confirm the hypothesis that the macrophage-secreted SHH is responsible for the increase we previously observed in the CSC population (Fig 1), we used siRNA to knock down SHH expression in RAW cells, thereby decreasing its secretion by macrophages (Fig 2C). The CM were collected from siSHH or control siRNA-transfected macrophages and incubated with CT26 or HCP-1 cells; the CSC population was analyzed using the sphere-forming assay. Fig 2D and 2E show that SHH knockdown dramatically decreased sphere formation in both cell lines (P < 0.01).
Next, we evaluated the effects of a small-molecule inhibitor of SMO, LDE225 ([47]. LDE225 significantly inhibited the sphere forming capacity in HCP-1 cells in a dose dependent manner following RAW + LPS CM and macrophage + LPS CM treatment (Fig 2F, P < 0.01). Taken together, these data confirmed that LPS-activated RAW macrophages, and human macrophage activated by LPS as well secrete SHH, which in turn, increases the CSC population in CRC cells.
Murine macrophage CM promotes chemoresistance of CRC cells
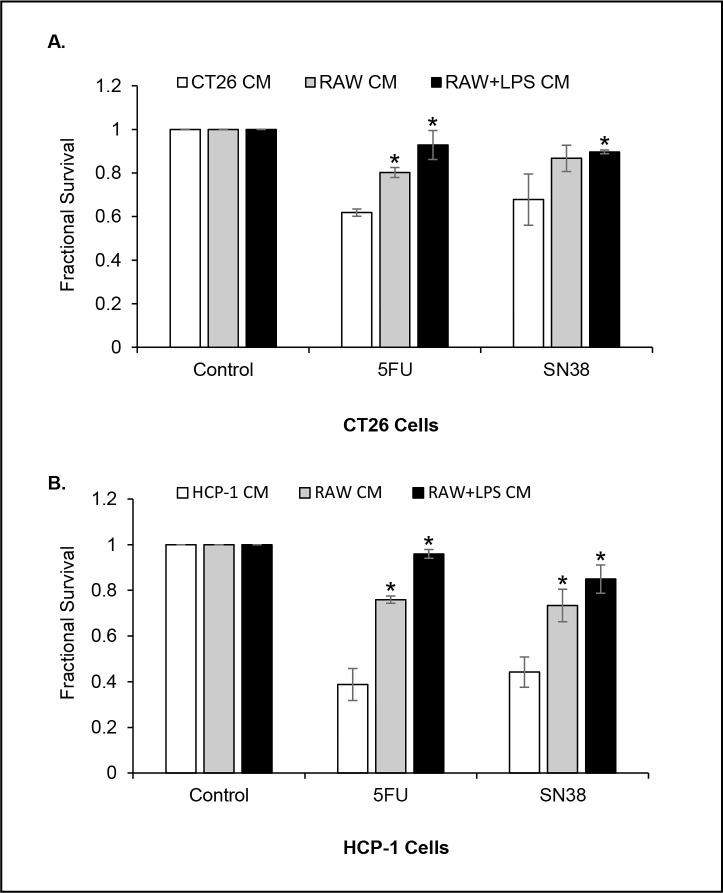
Preclinical studies suggested that CSCs possess intrinsic properties that mediate their resistance to chemotherapy [16, 17, 48]. Since our previous data showed that macrophage CM enhanced the CSC phenotype, we assessed the CRC cells response to chemotherapy after a 48 h pretreatment with murine macrophage CM. After pretreatment, the CRC cells were exposed to either 5-fluorouracil (5-FU, 2 μg/ml) or SN38 (20 nM) in LPS-activated macrophage CM or control CM for 72 h and cell survival was assessed by the MTT assay. Our results showed that both CRC cell lines grown in CT26 CM or HCP-1 CM exhibited a significantly higher sensitivity to 5FU or SN38, whereas, the cells cultured in LPS-activated macrophage CM demonstrated a significantly higher survival rate after 5-FU or SN38 treatment (Fig 3A and 3B, P < 0.05 vs CT26 CM; P < 0.001 vs HCP-1 CM, respectively). We also observed that non-activated macrophage CM induced resistance to 5FU and SN38 in CRC cells, suggesting that non-activated macrophages can also influence chemoresistance in tumor cells.
Fig 3. Murine macrophage CM promotes chemoresistance of CRC cells.
(A and B) CM pretreated CT26 or HCP-1 cells were incubated with 5-FU or SN38 in control CM or RAW+LPS CM and cell viability was determined by MTT assay (*P < 0.05; *P < 0.01 respectively).
Human macrophage CM promotes the self-renewal capacity of CRC cells
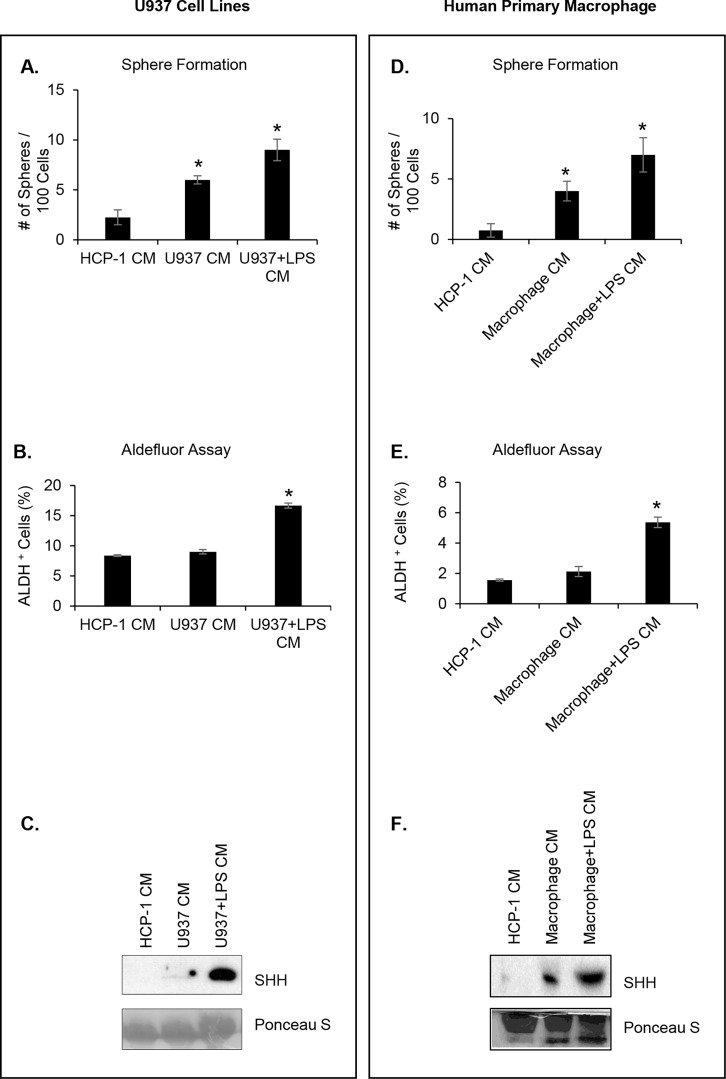
Finally, we confirmed the effect of macrophages CM on CSC using the human U937 monocyte cell line. U937 monocytes were polarized by LPS, and the population was characterized by RT-PCR (S1B Fig). As previously noted with RAW cells, we observed that LPS-activated U937 cells were a mixture of M1-like and M2-like macrophages.
The CM were prepared as previously described. After the HCP-1 cell line had been incubated with either U937 CM or LPS-activated U937 CM, the sphere formation assay showed a significant increase in HCP-1 sphere-forming capability. In particular, LPS-activated U937 CM increased the number of spheres by more than four-fold compared to control CM (Fig 4A, P < 0.001). In addition, LPS-activated U937 CM treatment enriched the Aldefluor-positive cell population of HCP-1 by two-fold compared to control CM (Fig 4B, P < 0.01). As expected, a Western blot analysis of the different CM showed that SHH was secreted at a higher rate by LPS-activated U937 cells compared to HCP-1 cells and U937 non-treated cells (Fig 4C). We also confirmed these results using human monocytes isolated from normal donor buffy coat. The results corroborated our previous findings: 1) the sphere formation assay showed a significantly increase in HCP-1 sphere-forming ability (6-fold) (Fig 4D; P < 0.01); 2) LPS-activated macrophage CM treatment enriched the Aldefluor-positive HCP-1 cell population by more than 3-fold compared to control CM (Fig 4E; P < 0.001); 3) Western blot analysis of the different CMs showed that SHH was secreted at a higher levels by LPS- activated macrophage compared to HCP-1 cells and untreated monocytes (Fig 4F).
Fig 4. Human macrophage CM promotes the self-renewal capacity of CRC cells.
(A) HCP-1 cells were cultured with LPS-activated U937 CM or control CM and the sphere forming assay was performed (*P < 0.001). (B) The Aldefluor-positive cell population was determined (*P < 0.01). (C) Western blot analysis of SHH secretion. Ponceau S served as loading control. (D) HCP-1 cells were cultured with LPS-activated human macrophage CM or control CM, sphere forming assay was performed (*P < 0.01). (E) The Aldefluor-positive cell population was determined (*P < 0.001). (F) Western blot analysis of SHH secretion. Ponceau S served as loading control.
Discussion
In the growing fields of immunotherapy and the tumor microenvironment, conflicting studies about the role of macrophages in tumor progression warrant continued investigation, especially in CRC. In this study, we examined the effect of classically LPS-activated macrophage CM on CRC cell lines, our results showed that LPS-activated macrophage CM mediates the CSC phenotype through activation of the SHH-Gli signaling pathway in CRC cells. Furthermore, we demonstrated that LPS-activated macrophage CM enhanced chemoresistance. Because macrophages display a broad functional spectrum and can change function depending on the microenvironment, their effects on tumor progression are difficult to predict. Such conflicts could be partly explained by the way macrophages are polarized by the tumor microenvironment [49]. Although TAMs were initially believed to be involved in mediating the pro-tumor activities of the M2 phenotype, it is now understood that they are probably composed of several distinct populations, with overlapping M1-like and M2-like features; these phenotypes may be dependent upon a variety of factors, including location in the microenvironment, stage of the tumor, and type of cancer [31, 50, 51]. Thus, the restrictive classification of TAMs simply as M1 or M2 does not accurately reflect their biological state in vivo. Our in vitro studies also demonstrated that LPS-activated macrophages were actually a mixture of both phenotypes (S1 Fig). This supports the hypothesis that macrophage activation leads to a mixed or double phenotype of M1 and M2 macrophages with populations likely ranging from one extreme to the other and also various phenotypes in between.
A growing body of evidence suggests that TAMs are critical for the self-renewal and maintenance of CSCs in established tumors [29–31]. Hedgehog (HH) is one of the major mediators involved in self-renewal and maintenance of CSCs [52]. The importance of this pathway has been demonstrated in various cancers including breast, lung, pancreas, colon and hepatic cancers [52]. Briefly, the HH pathway is activated by a family of ligands, three of which have been identified in humans: Sonic hedgehog (SHH), Indian hedgehog (IHH) and Desert hedgehog (DHH). Of these, SHH has been extensively studied [53]. Patched-1 (PTCH1), the receptor of SHH, inhibits Smoothened (SMO), a downstream protein in the HH pathway. However, upon SHH binding the inhibition of SHO by PTCH1 is relieved. These events lead to subsequent activation of the GLI transcription factors: the activators Gli1 and Gli2, and the repressor Gli3 [53]. In this study, we showed for the first time, that even classically activated macrophages are able to enrich the CSC population of CRC cell lines and that this enrichment is mediated by the SHH protein that is present in the macrophage CM. A previous study indicated that TAMs express milk-fat globule-epidermal growth factor-V111 (MFG-E8) that leads to enhanced pSTAT3 and SMO in CRC CSCs [35]. Increase in SMO in CSCs were linked to increased activity of the SHH pathway, an important regulator of stemness. Our observation that the LPS-activated macrophage CM itself contains high amounts of SHH and that treating CRC cells with this CM induces significant increases in GLI activity and increase in stem cell population, suggests a novel and more direct mechanism of activation of the SHH pathway. However, we have not been able to determine whether this increase in the relative CSC population was due to selection of CSCs or by direct stimulation of the CSC phenotype of CRC.
In summary, our findings emphasize the complexity of macrophage biology in the tumor microenvironment. Our results underline, 1) the ability of classically activated macrophages to stimulate the CSC phenotype in CRC cells, and, 2) LPS-activated macrophage CM promotes chemoresistance of CRC cells. Since macrophages are able to stimulate the CSC phenotype and promote chemoresistance, our results add to the consensus that macrophages are of critical importance to tumors response to treatment and that due to their plasticity can influence tumor growth and possibly, response to treatment. The multifactorial roles of macrophages are key in tumor-promoting or tumor-suppressing processes and should be considered overall in the design of anti-tumor therapeutics.
Supporting information
Characterization of the LPS-activated macrophage population through the detection of canonical M1 and M2 macrophage polarization markers in vitro.
(TIF)
Promoter activity of Hes-1, Gli, and TCF in HCP-1 cells after treatment with control CM or macrophage CM.
(TIF)
f: forward; r: reverse.
(TIF)
Acknowledgments
The authors thank Ann Sutton from the Department of Scientific Publications and Rita Hernandez from the Department of Surgical Oncology at The University of Texas MD Anderson Cancer Center for editorial assistance.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by NIH grant RO1 CA157880 to LME; DOD grant CA100879 to LME and the William C. Liedke, Jr., Chair in Cancer Research to LME. The funders had no role in study design, data collection analysis, decision to publish, or preparation of the manuscript.
References
- 1.Fakih MG. Metastatic Colorectal Cancer: Current State and Future Directions. J Clin Oncol. 2015;33(16):1809–24. doi: 10.1200/JCO.2014.59.7633 . [DOI] [PubMed] [Google Scholar]
- 2.Prenen H, Vecchione L, Van Cutsem E. Role of targeted agents in metastatic colorectal cancer. Target Oncol. 2013;8(2):83–96. Epub 2013/05/07. doi: 10.1007/s11523-013-0281-x . [DOI] [PubMed] [Google Scholar]
- 3.Davies JM, Goldberg RM. Treatment of metastatic colorectal cancer. Semin Oncol. 2011;38(4):552–60. Epub 2011/08/04. doi: 10.1053/j.seminoncol.2011.05.009 [pii]. . [DOI] [PubMed] [Google Scholar]
- 4.Dalerba P, Dylla SJ, Park IK, Liu R, Wang X, Cho RW, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci U S A. 2007;104(24):10158–63. doi: 10.1073/pnas.0703478104 ; PubMed Central PMCID: PMC1891215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1(5):555–67. doi: 10.1016/j.stem.2007.08.014 ; PubMed Central PMCID: PMC2423808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Storms RW, Trujillo AP, Springer JB, Shah L, Colvin OM, Ludeman SM, et al. Isolation of primitive human hematopoietic progenitors on the basis of aldehyde dehydrogenase activity. Proc Natl Acad Sci U S A. 1999;96(16):9118–23. ; PubMed Central PMCID: PMC17742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lohberger B, Rinner B, Stuendl N, Absenger M, Liegl-Atzwanger B, Walzer SM, et al. Aldehyde dehydrogenase 1, a potential marker for cancer stem cells in human sarcoma. PLoS One. 2012;7(8):e43664 doi: 10.1371/journal.pone.0043664 ; PubMed Central PMCID: PMC3426519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim MP, Fleming JB, Wang H, Abbruzzese JL, Choi W, Kopetz S, et al. ALDH activity selectively defines an enhanced tumor-initiating cell population relative to CD133 expression in human pancreatic adenocarcinoma. PLoS One. 2011;6(6):e20636 doi: 10.1371/journal.pone.0020636 ; PubMed Central PMCID: PMC3113804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaur P, Sceusi EL, Samuel S, Xia L, Fan F, Zhou Y, et al. Identification of cancer stem cells in human gastrointestinal carcinoid and neuroendocrine tumors. Gastroenterology. 2011;141(5):1728–37. doi: 10.1053/j.gastro.2011.07.037 ; PubMed Central PMCID: PMC3202668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cioffi M, D'Alterio C, Camerlingo R, Tirino V, Consales C, Riccio A, et al. Identification of a distinct population of CD133(+)CXCR4(+) cancer stem cells in ovarian cancer. Scientific reports. 2015;5:10357 doi: 10.1038/srep10357 ; PubMed Central PMCID: PMC4650662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desiderio V, Papagerakis P, Tirino V, Zheng L, Matossian M, Prince ME, et al. Increased fucosylation has a pivotal role in invasive and metastatic properties of head and neck cancer stem cells. Oncotarget. 2015;6(1):71–84. doi: 10.18632/oncotarget.2698 ; PubMed Central PMCID: PMC4381579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lobo NA, Shimono Y, Qian D, Clarke MF. The biology of cancer stem cells. Annual review of cell and developmental biology. 2007;23:675–99. doi: 10.1146/annurev.cellbio.22.010305.104154 . [DOI] [PubMed] [Google Scholar]
- 13.Tirino V, Desiderio V, Paino F, De Rosa A, Papaccio F, La Noce M, et al. Cancer stem cells in solid tumors: an overview and new approaches for their isolation and characterization. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2013;27(1):13–24. doi: 10.1096/fj.12-218222 . [DOI] [PubMed] [Google Scholar]
- 14.Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445(7123):111–5. doi: 10.1038/nature05384 . [DOI] [PubMed] [Google Scholar]
- 15.O'Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445(7123):106–10. doi: 10.1038/nature05372 . [DOI] [PubMed] [Google Scholar]
- 16.Al-Hajj M. Cancer stem cells and oncology therapeutics. Curr Opin Oncol. 2007;19(1):61–4. doi: 10.1097/CCO.0b013e328011a8d6 . [DOI] [PubMed] [Google Scholar]
- 17.Wicha MS, Liu S, Dontu G. Cancer stem cells: an old idea—a paradigm shift. Cancer research. 2006;66(4):1883–90; discussion 95–6. doi: 10.1158/0008-5472.CAN-05-3153 . [DOI] [PubMed] [Google Scholar]
- 18.Botchkina G. Colon cancer stem cells—from basic to clinical application. Cancer Lett. 2013;338(1):127–40. Epub 2012/04/28. doi: 10.1016/j.canlet.2012.04.006 [pii]. . [DOI] [PubMed] [Google Scholar]
- 19.Flemming A. Cancer stem cells: Targeting the root of cancer relapse. Nat Rev Drug Discov. 2015;14(3):165 Epub 2015/02/28. doi: 10.1038/nrd4560 [pii]. . [DOI] [PubMed] [Google Scholar]
- 20.Todaro M, Francipane MG, Medema JP, Stassi G. Colon cancer stem cells: promise of targeted therapy. Gastroenterology. 2010;138(6):2151–62. Epub 2010/04/28. doi: 10.1053/j.gastro.2009.12.063 [pii]. . [DOI] [PubMed] [Google Scholar]
- 21.Takebe N, Harris PJ, Warren RQ, Ivy SP. Targeting cancer stem cells by inhibiting Wnt, Notch, and Hedgehog pathways. Nat Rev Clin Oncol. 2011;8(2):97–106. Epub 2010/12/15. doi: 10.1038/nrclinonc.2010.196 [pii]. . [DOI] [PubMed] [Google Scholar]
- 22.Butler JM, Kobayashi H, Rafii S. Instructive role of the vascular niche in promoting tumour growth and tissue repair by angiocrine factors. Nat Rev Cancer. 2010;10(2):138–46. doi: 10.1038/nrc2791 ; PubMed Central PMCID: PMC2944775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosen JM, Jordan CT. The increasing complexity of the cancer stem cell paradigm. Science. 2009;324(5935):1670–3. doi: 10.1126/science.1171837 ; PubMed Central PMCID: PMC2873047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding J, Jin W, Chen C, Shao Z, Wu J. Tumor associated macrophage x cancer cell hybrids may acquire cancer stem cell properties in breast cancer. PLoS One. 2012;7(7):e41942 doi: 10.1371/journal.pone.0041942 ; PubMed Central PMCID: PMC3405038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Borovski T, De Sousa EMF, Vermeulen L, Medema JP. Cancer stem cell niche: the place to be. Cancer research. 2011;71(3):634–9. doi: 10.1158/0008-5472.CAN-10-3220 . [DOI] [PubMed] [Google Scholar]
- 26.Lu J, Ye X, Fan F, Xia L, Bhattacharya R, Bellister S, et al. Endothelial cells promote the colorectal cancer stem cell phenotype through a soluble form of Jagged-1. Cancer Cell. 2013;23(2):171–85. Epub 2013/02/05. doi: 10.1016/j.ccr.2012.12.021 ; PubMed Central PMCID: PMCPMC3574187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whiteside TL. The tumor microenvironment and its role in promoting tumor growth. Oncogene. 2008;27(45):5904–12. doi: 10.1038/onc.2008.271 ; PubMed Central PMCID: PMCPMC3689267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mantovani A. MSCs, macrophages, and cancer: a dangerous menage-a-trois. Cell Stem Cell. 2012;11(6):730–2. doi: 10.1016/j.stem.2012.11.016 . [DOI] [PubMed] [Google Scholar]
- 29.Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol. 2010;11(10):889–96. doi: 10.1038/ni.1937 . [DOI] [PubMed] [Google Scholar]
- 30.Raggi C, Mousa HS, Correnti M, Sica A, Invernizzi P. Cancer stem cells and tumor-associated macrophages: a roadmap for multitargeting strategies. Oncogene. 2015. doi: 10.1038/onc.2015.132 . [DOI] [PubMed] [Google Scholar]
- 31.Sainz B Jr., Carron E, Vallespinos M, Machado HL. Cancer Stem Cells and Macrophages: Implications in Tumor Biology and Therapeutic Strategies. Mediators Inflamm. 2016;2016:9012369 doi: 10.1155/2016/9012369 ; PubMed Central PMCID: PMCPMC4769767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitchem JB, Brennan DJ, Knolhoff BL, Belt BA, Zhu Y, Sanford DE, et al. Targeting tumor-infiltrating macrophages decreases tumor-initiating cells, relieves immunosuppression, and improves chemotherapeutic responses. Cancer research. 2013;73(3):1128–41. doi: 10.1158/0008-5472.CAN-12-2731 ; PubMed Central PMCID: PMC3563931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wan S, Zhao E, Kryczek I, Vatan L, Sadovskaya A, Ludema G, et al. Tumor-associated macrophages produce interleukin 6 and signal via STAT3 to promote expansion of human hepatocellular carcinoma stem cells. Gastroenterology. 2014;147(6):1393–404. doi: 10.1053/j.gastro.2014.08.039 ; PubMed Central PMCID: PMC4253315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fan QM, Jing YY, Yu GF, Kou XR, Ye F, Gao L, et al. Tumor-associated macrophages promote cancer stem cell-like properties via transforming growth factor-beta1-induced epithelial-mesenchymal transition in hepatocellular carcinoma. Cancer Lett. 2014;352(2):160–8. doi: 10.1016/j.canlet.2014.05.008 . [DOI] [PubMed] [Google Scholar]
- 35.Jinushi M, Chiba S, Yoshiyama H, Masutomi K, Kinoshita I, Dosaka-Akita H, et al. Tumor-associated macrophages regulate tumorigenicity and anticancer drug responses of cancer stem/initiating cells. Proc Natl Acad Sci U S A. 2011;108(30):12425–30. doi: 10.1073/pnas.1106645108 ; PubMed Central PMCID: PMC3145680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest. 2012;122(3):787–95. doi: 10.1172/JCI59643 ; PubMed Central PMCID: PMCPMC3287223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Braster R, Bogels M, Beelen RH, van Egmond M. The delicate balance of macrophages in colorectal cancer; their role in tumour development and therapeutic potential. Immunobiology. 2015. doi: 10.1016/j.imbio.2015.08.011 . [DOI] [PubMed] [Google Scholar]
- 38.Erreni M, Mantovani A, Allavena P. Tumor-associated Macrophages (TAM) and Inflammation in Colorectal Cancer. Cancer Microenviron. 2011;4(2):141–54. doi: 10.1007/s12307-010-0052-5 ; PubMed Central PMCID: PMC3170420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Biswas SK, Allavena P, Mantovani A. Tumor-associated macrophages: functional diversity, clinical significance, and open questions. Semin Immunopathol. 2013;35(5):585–600. doi: 10.1007/s00281-013-0367-7 . [DOI] [PubMed] [Google Scholar]
- 40.Fan F, Bellister S, Lu J, Ye X, Boulbes DR, Tozzi F, et al. The requirement for freshly isolated human colorectal cancer (CRC) cells in isolating CRC stem cells. British journal of cancer. 2015;112(3):539–46. doi: 10.1038/bjc.2014.620 ; PubMed Central PMCID: PMC4453647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377(6547):355–8. doi: 10.1038/377355a0 . [DOI] [PubMed] [Google Scholar]
- 42.Limgala R CY, Atsushi Terunuma, Philip Martin, Kathy Kelly, Jonathan Vogel, editor Isolation of Prostate Cancer Stem Cells using Developmental Signaling Pathway Activities. AACR Annual Meeting; 2009 Apr 18–22, 2009; Denver, CO.
- 43.Katoh M, Katoh M. Notch signaling in gastrointestinal tract (review). International journal of oncology. 2007;30(1):247–51. . [PubMed] [Google Scholar]
- 44.Vermeulen L, De Sousa EMF, van der Heijden M, Cameron K, de Jong JH, Borovski T, et al. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nature cell biology. 2010;12(5):468–76. doi: 10.1038/ncb2048 . [DOI] [PubMed] [Google Scholar]
- 45.Saif MW, Chu E. Biology of colorectal cancer. Cancer journal. 2010;16(3):196–201. doi: 10.1097/PPO.0b013e3181e076af . [DOI] [PubMed] [Google Scholar]
- 46.Marigo V, Roberts DJ, Lee SM, Tsukurov O, Levi T, Gastier JM, et al. Cloning, expression, and chromosomal location of SHH and IHH: two human homologues of the Drosophila segment polarity gene hedgehog. Genomics. 1995;28(1):44–51. . [DOI] [PubMed] [Google Scholar]
- 47.Della Corte CM, Bellevicine C, Vicidomini G, Vitagliano D, Malapelle U, Accardo M, et al. SMO Gene Amplification and Activation of the Hedgehog Pathway as Novel Mechanisms of Resistance to Anti-Epidermal Growth Factor Receptor Drugs in Human Lung Cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2015;21(20):4686–97. doi: 10.1158/1078-0432.CCR-14-3319 . [DOI] [PubMed] [Google Scholar]
- 48.Dallas NA, Xia L, Fan F, Gray MJ, Gaur P, van Buren G 2nd, et al. Chemoresistant colorectal cancer cells, the cancer stem cell phenotype, and increased sensitivity to insulin-like growth factor-I receptor inhibition. Cancer research. 2009;69(5):1951–7. doi: 10.1158/0008-5472.CAN-08-2023 ; PubMed Central PMCID: PMC3198868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cui YL, Li HK, Zhou HY, Zhang T, Li Q. Correlations of tumor-associated macrophage subtypes with liver metastases of colorectal cancer. Asian Pac J Cancer Prev. 2013;14(2):1003–7. . [DOI] [PubMed] [Google Scholar]
- 50.Mantovani A, Sozzani S, Locati M, Allavena P, Sica A. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002;23(11):549–55. . [DOI] [PubMed] [Google Scholar]
- 51.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8(12):958–69. doi: 10.1038/nri2448 ; PubMed Central PMCID: PMCPMC2724991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Della Corte CM, Viscardi G, Papaccio F, Esposito G, Martini G, Ciardiello D, et al. Implication of the Hedgehog pathway in hepatocellular carcinoma. World journal of gastroenterology. 2017;23(24):4330–40. doi: 10.3748/wjg.v23.i24.4330 ; PubMed Central PMCID: PMC5487497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nybakken K, Perrimon N. Hedgehog signal transduction: recent findings. Current opinion in genetics & development. 2002;12(5):503–11. . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Characterization of the LPS-activated macrophage population through the detection of canonical M1 and M2 macrophage polarization markers in vitro.
(TIF)
Promoter activity of Hes-1, Gli, and TCF in HCP-1 cells after treatment with control CM or macrophage CM.
(TIF)
f: forward; r: reverse.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.