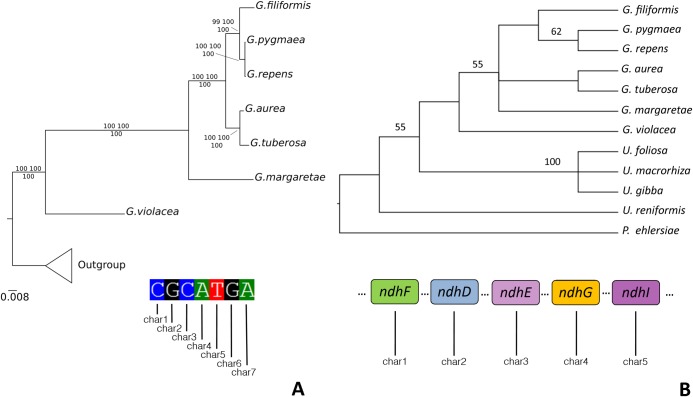
Fig 5. Phylogenetic hypothesis based on ndh sequences.
A. Analyses based on ndh sites (nucleotide-by-nucleotide). In this analysis, each nucleotide was used as one character (e.g. char1, char2, char3) B. Strict consensus of the two most parsimonious trees (33 steps; IC = 0.70; IR = 0.78) based on the matrix codified for ndh patterns. In this analysis, each ndh gene was applied to two characters: one codified as absent (state 0) or present (state 1) (characters 1 to 11) and other codified as pseudogenized (state 0), decayed (state 1), complete (state 2) and inapplicable (state “-”, when the gene is deleted) (characters 12 to 22). For details see S2, S3 and S4 Tables.