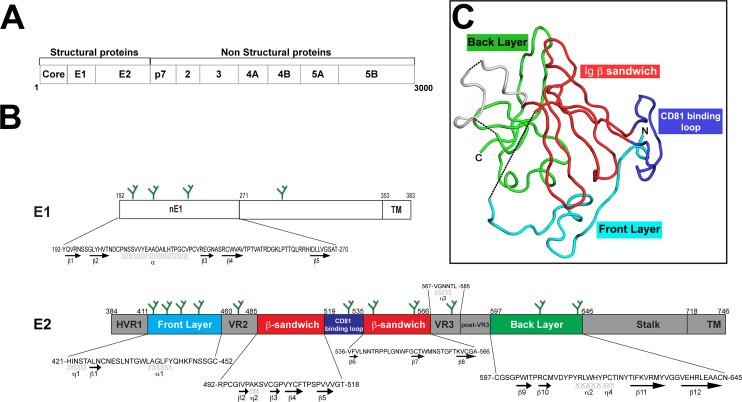
Fig 1. Overview of E1 and E2 glycoprotein structures.
(A) The approximately 3000 amino-acid HCV polyprotein generates 10 proteins following cleavage, of which E1 and E2 are two of the three structural proteins. (B) Spanning amino acids 192–383, the structure of E1 is poorly understood although crystallization of the first half of the protein (aa192-271) revealed secondary structures that could be present in native E1 [19] including an α-helix flanked by several β-sheets. In contrast, E2 (aa 384–746) has several well-defined regions containing β-sheets, α-helices, and η (310) helices. Sequences for regions in which secondary structure is known (e.g. nE1, E2 front layer, β-sandwich core, back layer, etc.) are included (prototypic H77 sequence). Green-branched forks depict relative locations of N-linked glycans. (C) Crystal structure of E2c [15] illustrated that E2 is characterized by a globular structure with a central Ig-like core flanked by front and back layers. The front layer, β-sandwich core, CD81 binding loop, and back layer are colored as depicted in panel (B).