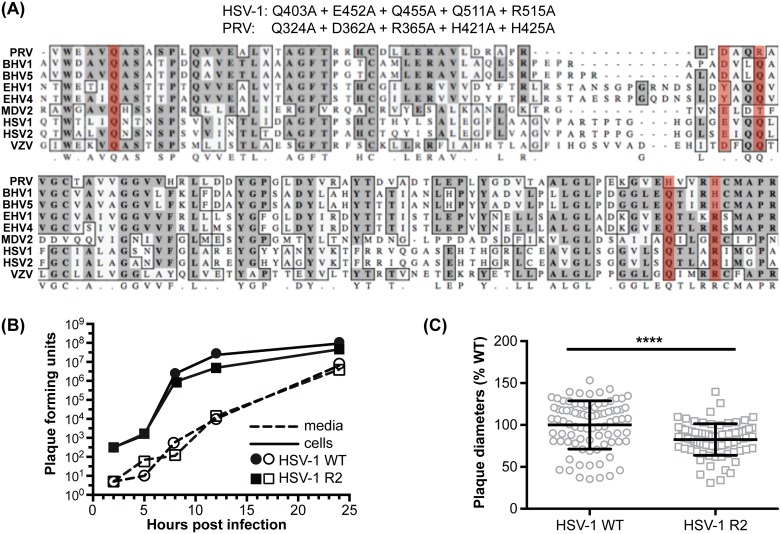
Fig 3. Mutation of the HSV-1 pUL37 R2 cluster phenocopies the PRV pUL37 R2 mutant.
(A) Amino acid mutations resulting from five codon changes introduced into the UL37 gene of HSV-1 are indicated at top, along with the corresponding mutations of the previously described PRV R2 mutant and the respective positions of the mutations within a sequence alignment of nine alpha-herpesviruses (mutated positions highlighted in red). (B) Wild-type (WT) and R2-mutant (R2) HSV-1 single-step propagation kinetics were determined by counting plaque-forming units harvested from media and Vero epithelial cells (cells) at the times indicated. Viruses used in this study were not modified to encode a fluorescent tag. (C) Fluorescent plaque diameters of WT and R2-mutant HSV-1 encoding mCherry fused to the pUL25 capsid protein were compiled from three independent experiments and plotted as a percentage of the average WT plaque diameter. Error bars are s.d. (****, p < 0.0001 based on a two-tailed unpaired t test).