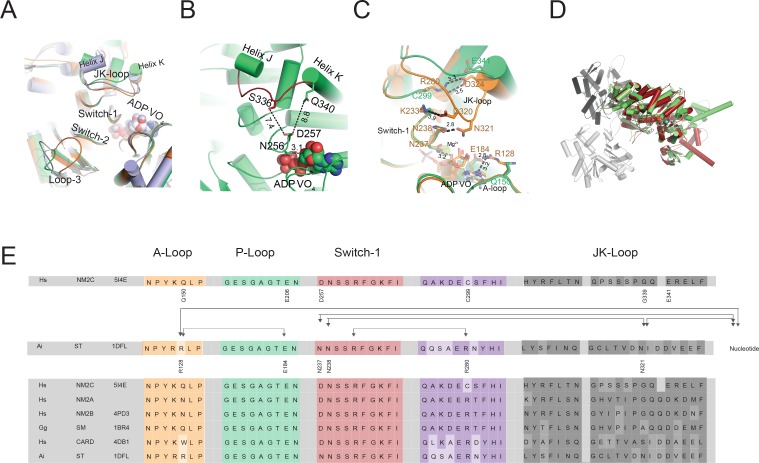
Figure 2. Conformational changes of the JK-loop in the myosin active site.
(A) Top view on the NM2C active site in the pre-powerstroke state (green) superimposed on pre-powerstroke state structures from chicken smooth muscle myosin-2 (grey, PDB entry 1BR4), Dictyostelium nonmuscle myosin-2 (blue, PDB entry 2XEL), and scallop striated muscle myosin-2 (orange, PDB entry 1QVI). The ATP analog ADP⋅VO4 is shown in spheres representation. (B) Conformation of the JK-loop in vicinity to the NM2C active site. The JK-loop flanks the active site and connects helices J and K. The distance between the residue Q340 of the JK-loop in the U50 kDa and the D257 of switch-1 of the active site is ~8.8 Å. The distance between residue S336 of the JK-loop and switch-1 D257 is ~7.4 Å. Switch-1 residue N256 interacts with α-phosphate (3.1 Å) and β-phosphate (3.5 Å) group of ADP⋅VO4 in the active site. NM2C is colored in green/orange, the JK-loop is colored in brick red and ADP⋅VO4 is shown in spheres. (C) Interactions between the JK-loop and the switch-1 region are compared between the NM2C (green) and scallop striated muscle myosin-2 (orange, PDB entry 1QVI). A-loop residue R128 is coordinating the interaction to the ADP adenosine in the active site of striated muscle myosin-2. The distance between the residues is 3.2 Å. R128 further forms a hydrogen bond (2.8 Å) with E184 of the P-loop. JK-loop N321 is in hydrogen bond interaction with switch-1 N238, located at a distance of 4.6 Å to the hydroxyl group of the C2’ of the ADP ribose. The connectivity between switch-1 and the nucleotide is further strengthened by a hydrogen bond between N237 and the ADP ribose. NM2C lacks all interactions described for scallop striated muscle myosin-2 due to the replacement of R128 with Q150 and JK-loop shortening which increases the distance to the adenosine in the active site to 5.8 Å and disrupts constrains between swich-1 and the JK-loop. All residues in the JK-loop region are labeled for scallop striated muscle myosin-2 (PDB entry 1QVI). For NM2C only amino acid substitutions are labeled for legibility. (D) Superimposition of the NM2C pre-powerstroke state structure (green) and the actin-bound near-rigor actoNM2C complex (red) shows that the nucleotide-binding site does not undergo major structural changes. Actin subunits are colored in shades of grey and the nucleotide is shown in spheres representation. (E) Sequence alignment of select structural elements in the myosin motor domain that interact with the JK-loop. Interactions of A-loop R128 are highlighted with brackets for scallop striated muscle myosin-2 (PDB entry 1QVI). All highlighted interactions are absent in NM2C due to the presence of Q150 in the A-loop. Abbreviations used: Hs NM2C: human NM2C (NP_079005.3); Hs NM2A: human nonmuscle myosin-2A (NP_002464.1); NM2B: human nonmuscle myosin-2B (NP_005955.3); Gg SM: chicken smooth muscle myosin-2 (NP_990605.2); Hs CARD: human beta β-cardiac muscle myosin-2 (NP_000248.2); Ai ST: scallop striated muscle myosin-2 (P24733.1). PDB entries are indicated when available.