Extended Data Figure 1. Substrate recruitment by the FRB domain.
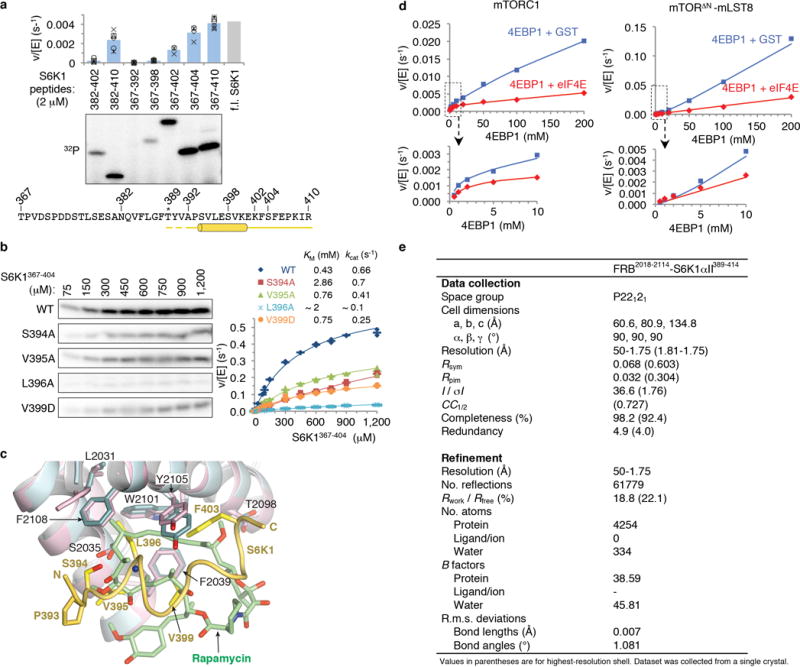
a, Deletion mapping of S6K1 FRB-binding motif polypeptides (2 μM) using phosphorylation by the mTORΔN-mLST8 (30 nM) as the assay, extending previous findings1. Truncation up-to 7 residues N-terminal to the Thr389 phosphorylation site (indicated by asterisk) has minimal effect, whereas C-terminal truncations starting 15 residues from Thr389 successively reduce phosphorylation. The polypeptides were produced as described in Methods for the S6K1367–404 peptide. Column graph shows velocity divided by enzyme concentration from the quantitation of the 32P autoradiogram. Columns show means and markers show values from independent experiments (n=6 for 382–410, 367–392, 367–398 and 367–410 reactions, n=5 for 382–402, n=4 for 367–404, and n=3 for 367–402). The column labeled f.l. S6K1 shows the phosphorylation level of full-length S6K1ki (ki superscript indicates the kinase-inactive K100R mutant) under the same conditions, as reported by Yang et al.1. Truncation of the S6K1 peptide to 20 residues (S6K1382–402), which is the standardized length used in the peptide library of Figure 1g, reduces phosphorylation to ~20 % of S6K1367–404, likely in part because of end-effects destabilizing the helix as well as eliminating some minor contacts. b, Michaelis-Menten steady state kinetic constants for mTORΔN-mLST8 (30 nM) phosphorylating wild type and the indicated mutant S6K1367–404 peptides, quantified as in a. Graph shows means (dashes) and values (markers as indicated) from independent experiments (n=3, except for the 1, 2, 10, 750, 900 and 1,200 μM points which are n=2). Also shown are the KM and kcat values, calculated by non-linear regression fitting of the data, above the graph, and their simulated curves in the graph. Mutations that significantly reduced phosphorylation but do not make substantial direct FRB contacts include S394A, which eliminates a hydrogen bond that stabilizes the helix N-terminus, and the helix breaking V395G mutation that further reduces phosphorylation compared to V395A (Fig. 1d); together, these point to the importance of the helical conformation. Additional mutations include Val391 and Pro393, in the segment between Thr389 and the start of the helix. Pro393 may be important for guiding the FRB-anchored substrate to the kinase active site, and that the P+2 residue Val391 may be involved in contacts to the kinase C lobe, where, by analogy to canonical kinases, the peptide segment of the phosphorylation site and its immediate vicinity are expected to bind to. c, Superposition of the FRB-S6K1 interface onto the FRB-rapamycin-FKBP12 structure2 (FKBP12 is omitted from clarity), highlighting the similarities in the binding of the Leu396 side chain to the same pocket as rapamycin’s key C23 methyl group and flanking portions of its triene arm. The FRB-S6K1 interface is colored as in Figure 1b, rapamycin green and its associated FRB domain cyan. In the crystals of the FRB-S6K1389–414 fusion protein, S6K1 residues 389–391 and 411–414 are disordered, while residues 405–410 are involved in crystal packing. d, Graph with the quantitation of the reactions shown in Figure 1h. e, X-ray data collection and refinement statistics for the FRB2018–2114-S6K1αII389–414 fusion protein structure.
