Figure 1. The FRB domain is a substrate-recruitment site.
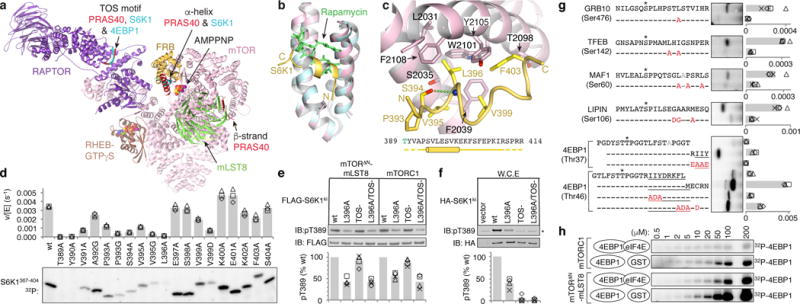
a, Composite image of S6K1 and PRAS40 sub-complex crystal structures superimposed on a RHEB-mTORC1 monomer from the cryo-EM structure. b, The FRB (pink) bound to the S6K1 peptide (yellow) superimposed on the FRB-Rapamycin-FKBP12 complex (blue FRB, green rapamycin; FKBP12 omitted). c, S6K1-FRB interface, S6K1 sequence and secondary structure (cylinder, helix; yellow dashes, unstructured). d, Phosphorylation of w.t. and mutant S6K1367–404 (2 μM) by mTORΔN-mLST8 (30 nM). 32P incorporation plotted as reaction velocity over enzyme concentration (means as columns, three independent experiments with markers). e, In vitro phosphorylation of w.t. and mutant FLAG-S6K1ki (2 μM) by 30 nM mTORΔN-mLST8 or mTORC1. f, Phosphorylation of w.t. and mutant HA-S6K1ki in HEK293 cells (asterisk, endogenous S6K1). In e and f, products detected by pT389-specific antibody and plotted relative to the w.t. reaction of each series (means as columns, four independent experiments with markers). g, Phosphorylation of indicated w.t. and mutant (red mutation) peptides (10 μM) of indicated substrates. Plotted as bars for means and markers for independent experiments (4EBP1, n=3; rest, n=4). 4EBP1 amphipathic helix underlined; asterisk, phosphorylation sites; gray, alanine mutated second phosphorylation sites. h, Phosphorylation of indicated 4EBP1 concentrations by 30 nM mTORΔN-mLST8 or mTORC1 in the presence of 1 molar equivalent eIF4E or GST control (plotted in Extended Data Fig. 1d).
