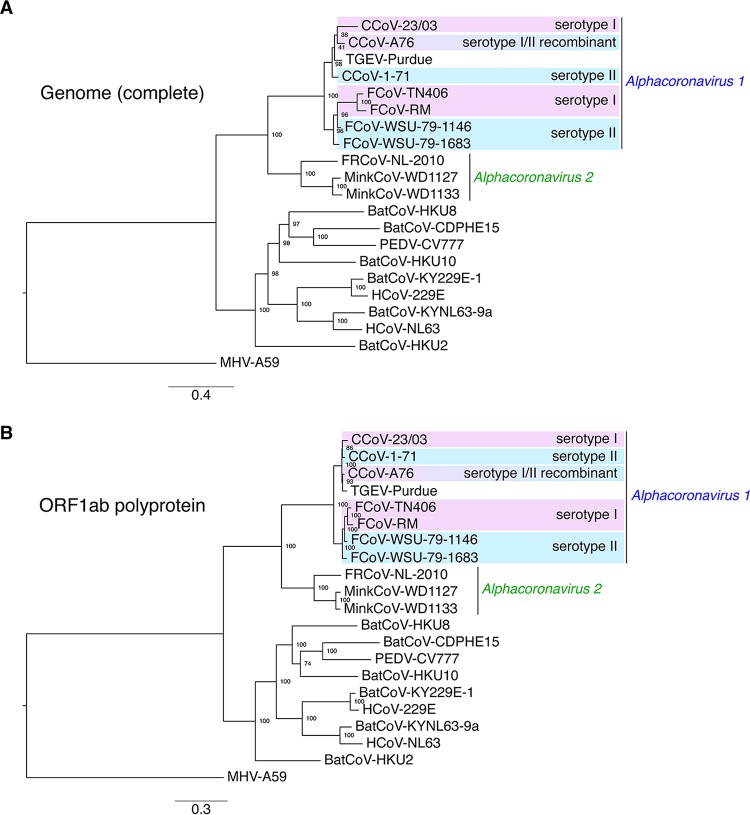
FIG 1 .
Phylogenetic analyses of alphacoronaviruses based on complete genome and ORF1ab protein sequence. Nucleotide sequences of the complete genomes of alphacoronaviruses and of the betacoronavirus MHV-A59 (A) or the complete protein sequences of the ORF1ab polyprotein of the corresponding viruses (B) were aligned using MAFFT within the Geneious 10 software package. The alignments were then used to generate maximum-likelihood phylogenetic trees using PhyML (25). The trees were rooted with MHV-A59. Numbers at nodes indicate the bootstrap support on 100 replicates. The scale bar indicates the estimated number of substitutions per site. Accession numbers for complete genome nucleotide sequences and ORF1ab protein sequences used are found in Materials and Methods.