FIG 4 .
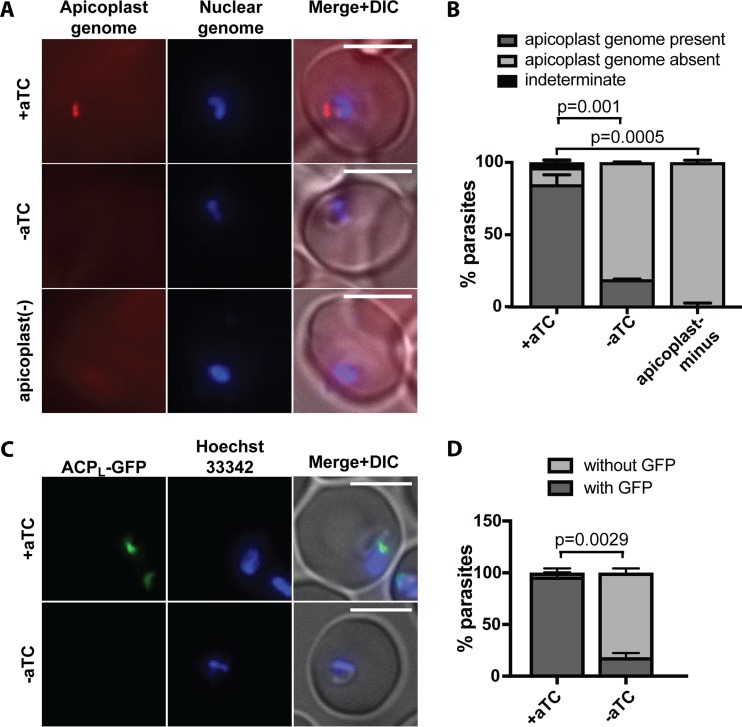
ATG8 knockdown leads to defects in apicoplast inheritance. (A) Representative images of apicoplast FISH detecting the apicoplast genome in ring-stage ATG8+ and ATG8-deficient parasites 48 h after aTC removal, i.e., after first reinvasion (top and middle rows, respectively). Apicoplast-minus parasites, generated by 4 cycles of chloramphenicol treatment and IPP rescue, are shown as a negative control for FISH staining (bottom row). Bars, 5 µm. (B) Quantification of parasites with or without apicoplast genome grown under indicated conditions. Mean ± standard deviation for 2 independent experiments as in panel A is shown. The P values, determined by one-way analysis of variance, are shown for the “apicoplast genome present” category. Data for separate replicates are in Fig. S4. (C) Representative images of ring-stage ATG8+ or ATG8-deficient parasites expressing ACPL-GFP 48 h after aTC removal, i.e., after first reinvasion. Bars, 5 µm. (D) Quantification of parasites with or without a discrete GFP-labeled structure as shown in panel C grown under indicated conditions. Mean ± standard deviation for 2 independent experiments is shown. The P value was determined by unpaired two-tailed t test for the “with GFP” category. Data for separate replicates are shown in Fig. S4. DIC, differential interference contrast.