Table 1. Data collection and refinement statistics of matrix protein 1 (M1) mutant structures.
| Data collection statistics | M(NLS-88R)-acidic | M(NLS-88R)-neutral | M(NLS-88E)-acidic | M(NLS-88E)-neutral |
|---|---|---|---|---|
| Final crystallization drop pH | pH 5.5 | pH 7.3 | pH 6.2 | pH 7.0 |
| Space group | P21 | P1 | P21212 | P21 |
| Cell dimensions (Å) | a=39.75, b=119.82, c=59.68; β=90.2° | a=27.7, b=33.3, c=36.2; α=112.2°, β=100.4°, γ=94.2° | a=85.61, b=133.26, c=39.31 | a=40.19, b=96.17, c=48.49; β=100.9° |
| Resolution (Å) | 29.95–2.0 (2.07–2.0) | 28.70–3.0 (3.11–3.00) | 26.23–2.5 (2.59–2.5) | 27.91–2.5 (2.59–2.5) |
| Measured reflections | 205147 | 6194 | 79188 | 44337 |
| Unique reflections | 33892 (3432) | 2187 (247) | 14742 (1473) | 12404 (1227) |
| Redundancy | 6.05 (5.97) | 2.83 (2.97) | 5.37 (5.51) | 3.57 (3.5) |
| I/σI | 13.1 (5.9) | 10.9 (1.5) | 12.1 (3.6) | 9.5 (2.7) |
| Completeness (%) | 90.0 (91.9) | 94.5 (93.2) | 90.7 (91.9) | 98.7 (99.3) |
| Rmerge (%)a | 8.8 (27.8) | 8.1 (51.2) | 7.7 (44.9) | 7.2 (37.9) |
| Structure refinement | ||||
| Resolution limit (Å) | 29.84–2.00 (2.07–2.0) | 21.93–3.00 (3.23–3.00) | 26.23–2.50 (2.69–2.50) | 27.58–2.50 (2.59–2.50) |
| No. of reflections | 33843 (3428) | 2177 (427) | 14714 (1470) | 12376 (1172) |
| Rwork (%) | 19.2 (24.0) | 27.2 (39.7) | 22.0 (30.2) | 22.8 (34.3) |
| Rfree (%)b | 23.6 (30.5) | 32.1 (43.4) | 31.2 (41.7) | 30.8 (31.0) |
| R.m.s.d. geometry | ||||
| Bond lengths (Å) | 0.006 | 0.003 | 0.009 | 0.008 |
| Bond angles | 1.1° | 0.595° | 1.165° | 1.3° |
| Dihedral angles (%) | ||||
| Most favored | 96.46 | 93.66 | 92.22 | 92.83 |
| Allowed regions | 2.09 | 5.63 | 5.83 | 2.93 |
| Av. B-factors (Å2) | ||||
| All atoms | 26.10 | 80.20 | 56.90 | 61.00 |
| Protein alone | 24.10 | 80.20 | 56.80 | 61.00 |
| Water | 36.80 | 69.20 | 47.50 | 58.00 |
| PDB ID | 5V6G | 5V8A | 5V7S | 5V7B |
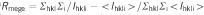 .
.
Rfree was calculated with 5% excluded reflection from the refinement.
