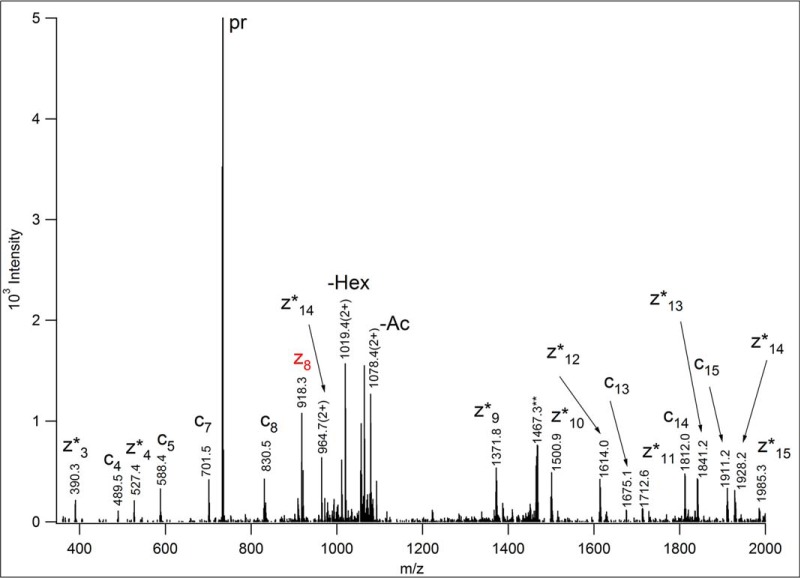
Fig. 6.
ETD spectrum of m/z 733.692(3+), acquired in the linear ion trap of an LTQ-Orbitrap Elite mass spectrometer (Thermo Fisher Scientific). AQGSQVLES(HexNAcHex)TPPPHVMR [682–698] of human ITIH2 protein (Uniprot ID: P19823) was identified from these data. In order to identify the peptide modified, common O-glycan structures were listed as variable modifications in the database search. The identity or the linkage of the glycans cannot be established from these data. The fragments were measured with low resolution, thus, the charge state of the ions was “determined” from the potential matches. Asterisks indicate the products of hydrogen migration, i.e. z+1 fragments. Although Ser-4 easily could be excluded as the glycosylation site, the C-terminal fragment printed in red provided the decisive information for the assignment of Ser-9 for that role, because it indicates that Thr-10 is unmodified. Some carbohydrate fragmentation was also detected: the loss of 42 Da from the N-acetylhexosamine, and some terminal hexose loss, both from the charge-reduced precursor ion. The precursor ion is labeled as “pr.” The **-labeled ion is the charge-reduced ion of a coeluting (2+) component.