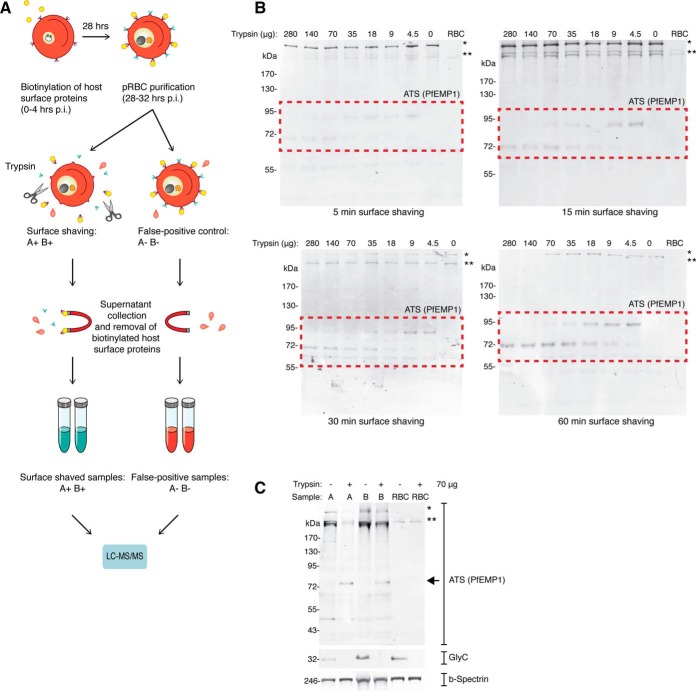
Fig. 2.
Supernatant proteomics. A, Overview of supernatant proteomics process. B, Surface shaving optimization. Intact and live trophozoite stage pRBC were surface shaved with a titration of trypsin, and analyzed by Western blot. Each lane represents protein extract from 2.5 × 106 pRBC. RBC refers to unparasitized RBCs. C, Validation of proteomics samples. After supernatants were collected for surface proteomics, the remaining cell pellet for each of the four samples were collected for Western blot analysis (A−, A+, B−, and B+). To validate the surface shaving method, membranes were probed with anti-Glycophorin C and anti-ATS antibodies. Anti-Spectrin antibodies were used as a loading control. Each lane represents protein extract from 2.5 × 106 pRBC.