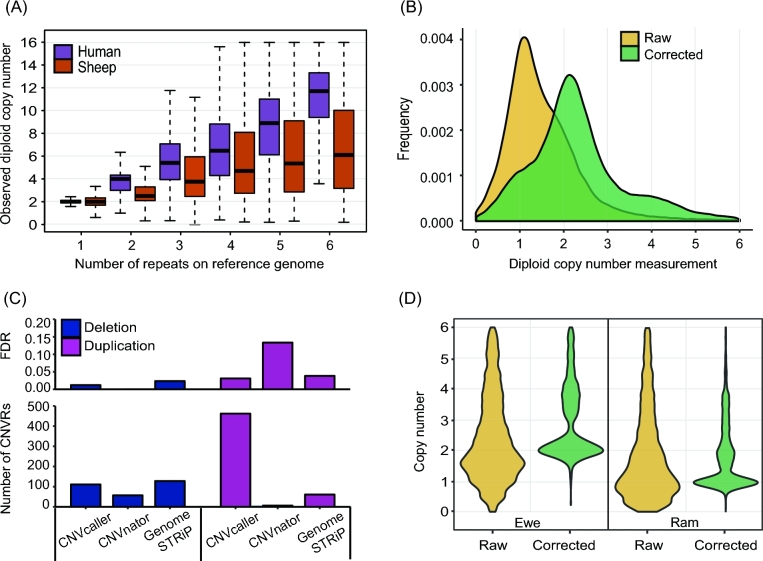
Figure 3:
Absolute copy number correction in the sheep genome. (A) The copy numbers of all the windows with no more than 6 repeats were plotted against the repeat numbers in the reference genome. Compared with humans, the sheep sample had much lower copy numbers in the putative duplicated regions than expected. (B) The distribution of copy numbers of the putative 2-copy regions in the sheep genome before and after absolute copy number correction. After correction, the main peak of the copy number shifted to 2 (normal diploid copy number). The smaller peaks at 4, after correction, indicated the 20% real SDs. (C) The number and FDR (Mendelian inconsistency) of detected CNVRs residing in the SD regions. The sheep SD regions include the regions longer than 2 kb with >97% identity. The CNVRs residing in the SD regions were defined if more than 50% of a given CNVR overlapped with the SD regions. (D) The raw and corrected copy numbers of all the X-linked scaffolds of 133 sheep.