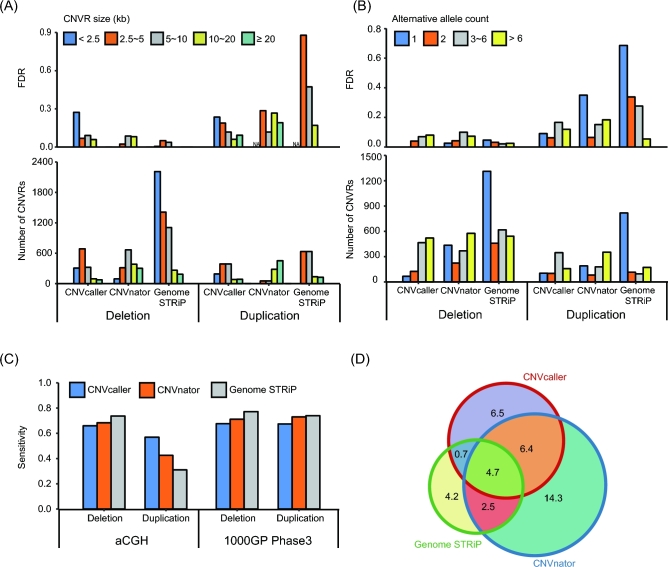
Figure 6:
Performance evaluations on the 1000GP data. (A, B) The number of calls (A) and the IRS FDR (B) partitioned by CNV length. All the calls on the autosomes were included. (C, D) The number of calls (C) and the IRS FDR (D) partitioned by allele frequency. To eliminate the huge FDR diversity of the short CNVs, the effect of the allele frequency was evaluated using the >2500 bp calls. (C) The sensitivity (the proportion of high-confidence CNV database overlapped by the predicted CNVs) of the 3 methods. For the highly variable FDRs, the sensitivity estimation removed the calls of less than 2500 bp or that had an alternative allele frequency of less than 5%. (D) Comparison of CNVR results identified by CNVcaller, CNVnator, and Genome STRiP based on the same 30 BAM files from the 1000GP Phase 3. The length of overlapping CNVRs was indicated in Mb.