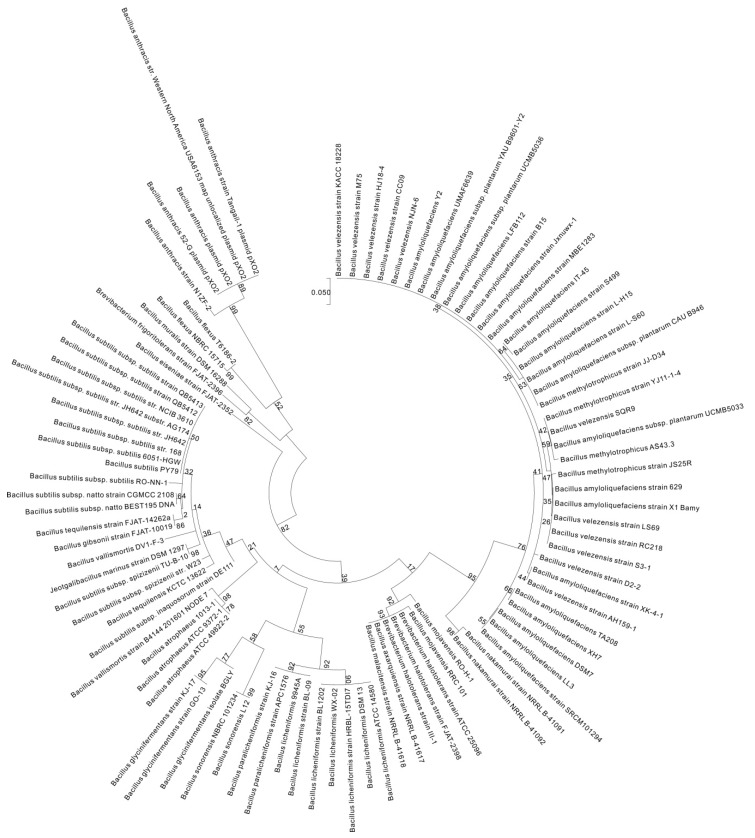
Figure 5.
Molecular phylogenetic analysis of pgsA by the maximum likelihood method. The phylogenetic tree with branch lengths was measured as the number of substitutions per site. The analysis involved 192 nucleotide sequences. Partial deletion was used and all positions with less than 95% site coverage were eliminated, i.e., less than 5% of the alignment gaps, instances of missing data, and ambiguous bases were allowed at any position. Scale bar: number of substitutions per site. Evolutionary analyses were conducted in MEGA7 [38].