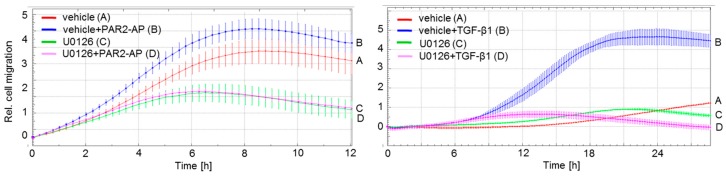
Figure 1.
PAR2–AP- and TGF-β1-driven random cell migration are dependent on ERK activation. Panc1 cells (left-hand graph) and Colo357 cells (right-hand graph) were subjected to a cell migration assay (chemokinesis setup) in the presence of vehicle (0.1% dimethylsulfoxide, DMSO) or 20 µM U0126 and either PAR2–AP (15 µM 2-furoyl-LIGRLO-NH2 (2f-LI), left-hand graph) or TGF-β1 (5 ng/mL, right-hand graph). In the left graph, differences are significant (p < 0.05, unpaired Student’s t-test) between vehicle + PAR2–AP treated cells (blue curve, tracing B) and U0126 + PAR2–AP treated cells (magenta curve, tracing D) at 4:00 and all later time points, and between vehicle treated cells (red curve, tracing A) and U0126 treated cells (green curve, tracing C) at 6:00 and all later time points. In the right-hand graph, differences are significant between vehicle + TGF-β1 treated cells (blue curve, tracing B) and U0126 + TGF-β1 treated cells (magenta curve, tracing D) at 12:00 and all later time points. For a color-independent identification of the curves, see letters to the right of each graph. In each graph, data are shown from one representative experiment; three experiments were performed in total.