Figure 2. Focus on technical factors using high-quality RNA probed with state-of-the-art miRNA microarrays.
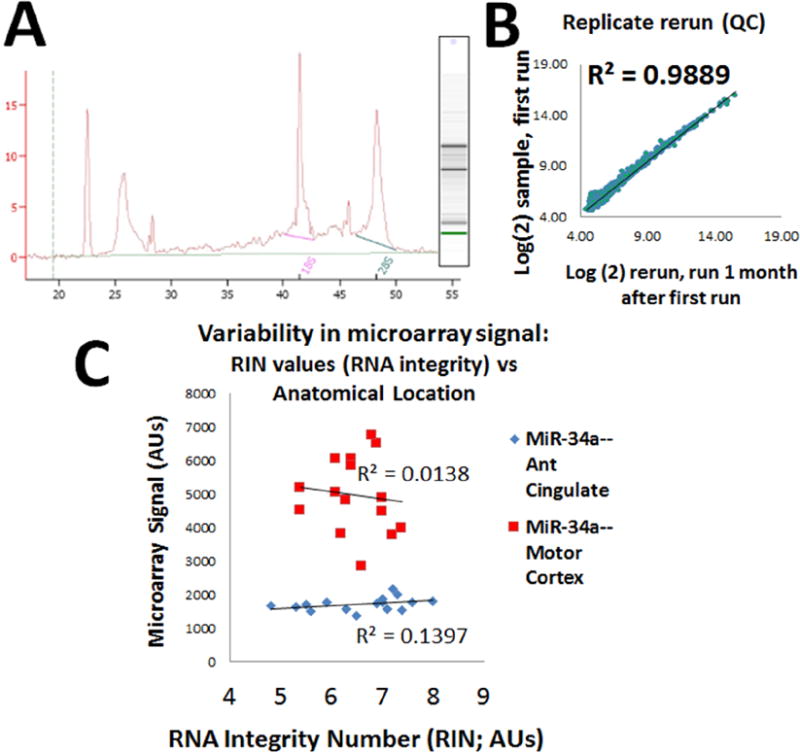
Quality control (QC) included Agilent BioAnalyzer (electropheragram, A), and the RNA Integrity Number (RIN) were mostly between 6-8, with inclusion criteria of RIN >4.0. Gray matter dissection was performed as described previously (Nelson et al., 2008b; Nelson and Wang, 2010; Wang et al., 2008c; Wang et al., 2011). For added QC, we performed extra technical replicates of six different samples a month after the primary microarrays were run and these showed within-case correlations (B is a representative correlation), demonstrating outstanding replicability. C. Variability in microarray signals correlated poorly with RIN number indicating that RNA integrity (>=4) and RNA degradation had weak impact on miRNA expression results in our preliminary samples. Shown here are results for miR-34a which are representative.
