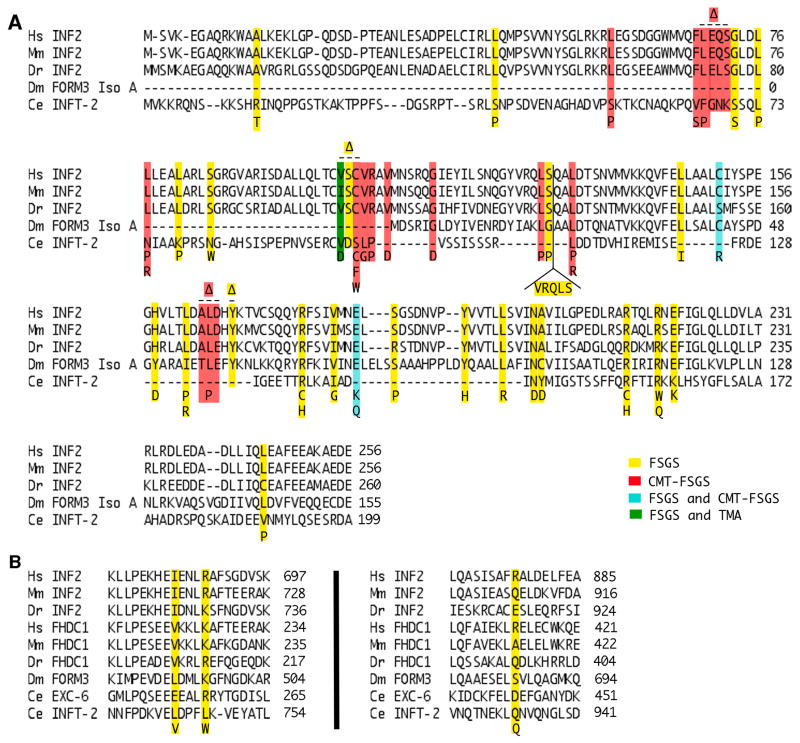
Figure 5. Conservation of disease-linked amino acids of INF2.
Shown are alignments of (A) DID or (B) FH2 domain sequences of inverted formins from human (Homo sapiens, Hs), mouse (Mus musculus, Mm), zebrafish (Danio rerio, Dr), fruit fly (Drosophila melanogaster, Dm), and worm (Caenorhabditis elegans, Ce). Disease-associated amino acid substitutions and insertions of human INF2 are shown below the alignment, while deletions ((x2206)) are shown above the alignment. Mutations are color coded to indicate association with FSGS (yellow), CMT-FSGS (red), both FSGS and CMT-FSGS (cyan), or both FSGS and TMA (green) [Brown et al., 2010; Boyer et al., 2011a; Boyer et al., 2011b; Lee et al., 2011; Gbadegesin et al., 2012; Barua et al., 2013; Lipska et al., 2013; Mademan et al., 2013; Rodriguez et al., 2013; Sanchez-Ares et al., 2013; Toyota et al., 2013; Caridi et al., 2014; Laurin et al., 2014; Park et al., 2014; Quaglia et al., 2014; Roos et al., 2015; Xie et al., 2015; Bullich et al., 2015; Jin et al., 2015; Münch et al., 2016; Rood et al., 2016; Challis et al., 2017]. Numbers indicate amino acid positions.