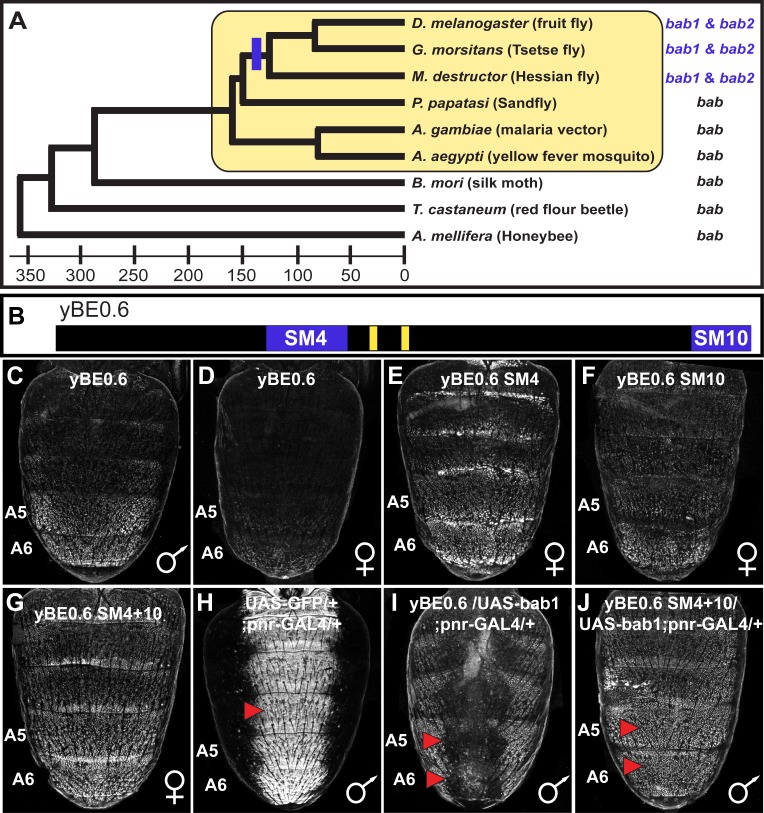
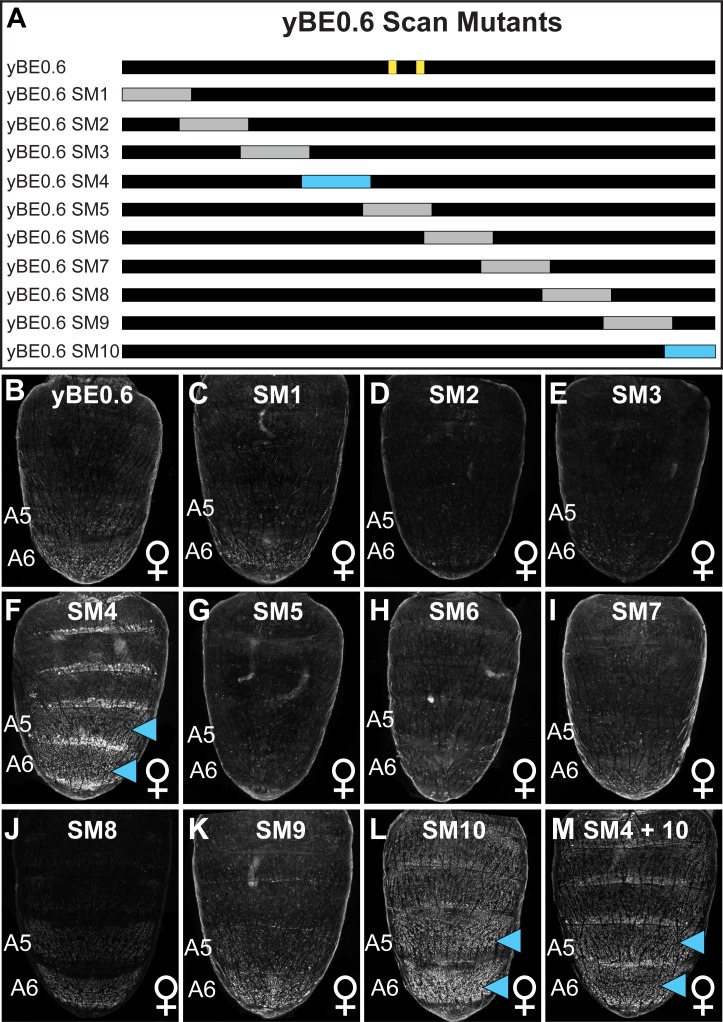
Figure 2. The tandemly duplicated bab genes perform a derived role in repressing a CRE controlling male-specific expression of the gene yellow.
(A) An ancestral bab gene was duplicated into the paralogous bab1 and bab2 genes in a Dipteran lineage that includes Drosophila fruit flies. The time scale indicates approximate divergence times in millions of years ago. (B) Male-specific expression of yellow in the abdominal epidermis is under the control of the yBE0.6 CRE that possesses two binding sites for Abd-B that are shown as yellow rectangles. Blue bars delimit the SM4 and SM10 regions required to suppress CRE activity in females. (C and D) The yBE0.6 EGFP reporter transgene is elevated in the male A5 and A6 abdomen segments (C) but is only barely detected females (D). (E–G) Ectopic reporter expression occurs in the female abdomen when either the SM4, SM10, or both regions are mutated. (H) The pnr-GAL4 driver activates dorsal midline expression of the UAS-EGFP gene, demarcating its domain of misexpression. (I) Dorsal midline expression of the yBE0.6 CRE is lost when bab1 is ectopically expressed by pnr-GAL4. (J) When the SM4 and SM10 regions are mutated, the yBE0.6 CRE can activate reporter expression in midline regions in spite of ectopically expressed bab1.