Abstract
Adaptation to air breathing after birth is dependent upon the synthesis and secretion of pulmonary surfactant by alveolar type 2 (AT2) cells. Surfactant, a complex mixture of phospholipids and proteins, is secreted into the alveolus, where it reduces collapsing forces at the air-liquid interface to maintain lung volumes during the ventilatory cycle. ABCA3, an ATP-dependent Walker domain containing transport protein, is required for surfactant synthesis and lung function at birth. Mutations in ABCA3 cause severe surfactant deficiency and respiratory failure in newborn infants. We conditionally deleted the Abca3 gene in AT2 cells in the mature mouse lung. Loss of ABCA3 caused alveolar cell injury and respiratory failure. ABCA3-related lung dysfunction was associated with surfactant deficiency, inflammation, and alveolar-capillary leak. Extensive but incomplete deletion of ABCA3 caused alveolar injury and inflammation, and it initiated proliferation of progenitor cells, restoring ABCA3 expression, lung structure, and function. M2-like macrophages were recruited to sites of AT2 cell proliferation during the regenerative process and were present in lung tissue from patients with severe lung disease caused by mutations in ABCA3. The remarkable and selective regeneration of ABCA3-sufficient AT2 progenitor cells provides plausible approaches for future correction of ABCA3 and other genetic disorders associated with surfactant deficiency and acute interstitial lung disease.
Keywords: Pulmonology
Keywords: Pulmonary surfactants
Surfactant deficiency, diffuse alveolar damage, and respiratory failure was followed by remarkable proliferation of alveolar cells and selective survival of ABCA3 sufficient cells.
Introduction
ABCA3, located on human chromosome 16p13.3, encodes a 1,704–amino acid (190-kDA), multimembrane-spanning protein expressed at high levels in AT2 cells in the lung (1–4). ABCA3 is required for packaging and secretion of surfactant lipids and is required for lung function at birth (5, 6). Autosomal recessive mutations in ABCA3 cause severe lung disease in infants and children, and they represent the most common genetic cause of respiratory failure in newborns (1, 7–9). ABCA3-related lung disease in infants is accompanied by lung injury and extensive tissue remodeling, leading to loss of alveolar structures that is generally fatal despite intensive care and ventilatory support (7, 10, 11). At present, lung transplantation is the only effective treatment for infants with severe ABCA3-related lung disease (12, 13).
The adult human lung consists of about 480 million alveoli, which are lined by epithelial alveolar type 1 and 2 (AT1 and AT2) cells (14). AT1 cells are large, squamous epithelial cells that are in close association with pulmonary endothelial cells, creating the extensive gas-exchange surface required for efficient exchange of oxygen and carbon dioxide after birth. AT2 cells comprise approximately 5% of the alveolar surface, are the sole source of pulmonary surfactant, and serve as the primary progenitors repairing the alveolar epithelium after injury (1, 15, 16). Pulmonary surfactant lipids and proteins are secreted into the alveoli — reducing surface tension at the air-liquid interface — and are required for lung function at birth and throughout life (1).
In the present study, we produced mice in which the Abca3 gene was selectively deleted in AT2 cells in the postnatal lung. Extensive loss of Abca3 resulted in respiratory failure and death caused by surfactant deficiency, alveolar-capillary leak, and inflammation consistent with the requirement of ABCA3 for lung function in newborn infants. Extensive but nonlethal deletion of Abca3 caused lung injury and inflammation, and it initiated alveolar cell proliferation that was followed by remarkable regeneration of Abca3-sufficient cells and restoration of lung structure and function.
Results
Deletion of Abca3 in AT2 cells caused respiratory failure.
Abca3flox/flox mice were designed to delete exons 4–7 under control of Cre-recombinase (17). To identify the role of Abca3 in postnatal lungs, transgenic Sftpc-CreERT2, Abca3flox/flox (Sftpc-CreERT2;Abca3flox/flox) (18) mice were produced, hereafter termed control. The Abca3 gene locus was selectively mutated in adult AT2 cells when control mice were treated with tamoxifen, hereafter termed Abca3–conditional KO (Abca3-cKO). Abca3 mRNA was decreased by 60% in whole lung from Abca3-cKO after 7 days (Figure 1A). Since ABCA3 is expressed in macrophages and other non-AT2 cells, the extent of Abca3 deletion was assessed in purified AT2 cells in which Abca3 mRNA was reduced by 90% (Figure 1B). Although Sftpc-CreERT2 expresses some active Cre in the absence of tamoxifen (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.97381DS1) (18), Abca3 mRNA was not altered in control mice (Abca3flox/flox 1.02 ± 0.18 vs. control 1.21 ± 0.54, P = 0.6; Supplemental Figure 2). To control for spontaneous recombination, untreated control mice were used. While some mutations in ABCA3 alter the distribution of surfactant proteins (5, 8), expression and processing of surfactant protein B (SP-B) and SP-C were unaltered in Abca3-cKO mice (Supplemental Figure 3).
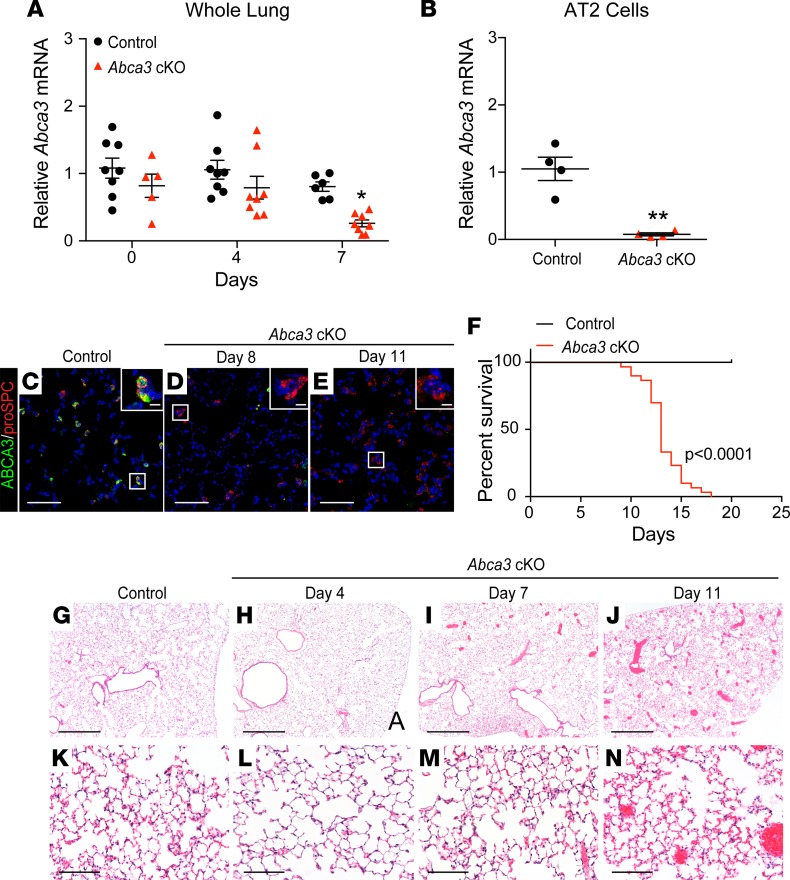
Figure 1. Deletion of Abca3 in AT2 cells causes respiratory failure.
(A) Quantitative PCR of Abca3 mRNA in whole lungs from adult control (black circles) and Abca3-cKO (red triangles) mice. (B) Abca3 mRNA in purified AT2 cells following 6 days of tamoxifen. Abca3 probes for exon 5–6 (A) and exon 3–4 (B), normalized to β-actin. Mean ± SEM, **P < 0.001, *P < 0.02 as determined by 1-way ANOVA, n = 4–8/group. Confocal immunofluorescence staining for ABCA3 (green) and proSPC (red) in control (C) and Abca3-cKO mice after 8 (D) and 11 days of tamoxifen (E). Scale bars: 50 μm. Inset cell scale bars: 5 μm. (F) Kaplan-Meier curve and statistical analysis (Wilcoxon-Gehan test) for control (black, n = 16) and Abca3-cKO (red, n = 30) mice, P < 0.0001. Representative lung histology of control (G and K) and Abca3-cKO mice after 4 (H and L), 7 (I and M), and 11 days of tamoxifen (J and N). Scale bars: 500 μm (G, H, I, and J) and 100 μm (K, L, M, and N), n = 3–4/group.
After exposure to tamoxifen, alveolar staining of ABCA3 was remarkably decreased, consistent with the loss of Abca3 mRNA (Figure 1, C–E). Abca3-cKO mice developed pulmonary inflammation, hyperemia, and alveolar wall thickening (Figure 1, G–N, and Supplemental Figure 4) accompanied by signs of respiratory failure, including dehydration, dyspnea, hunched appearance, and lethargy requiring euthanasia or causing death 12–15 days after tamoxifen (Figure 1F). Lung histology and analysis of bronchoalveolar lavage fluid (BALF) demonstrated mild lung inflammation (Figure 2, A and B) with increased numbers of neutrophils at later time points.
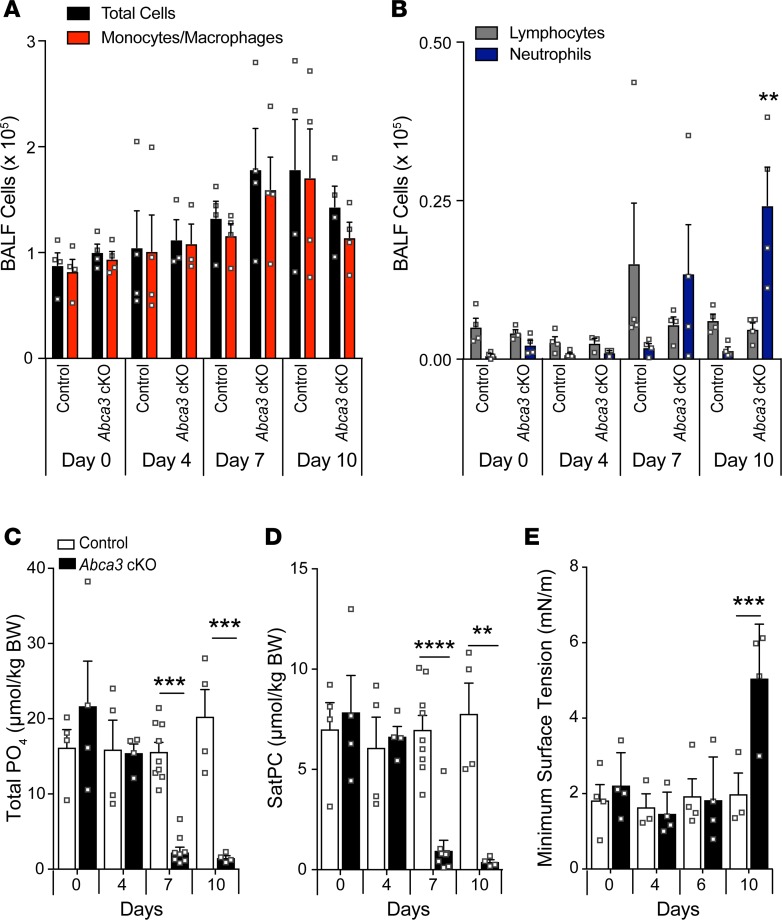
Figure 2. Inflammation and decreased lung phospholipids after deletion of Abca3.
(A–B) Differential cell numbers were determined in BALF by Diff-Quik staining. (C) Total lung phospholipid and saturated phosphatidylcholine (SatPC; D) levels were measured in BALF, and the function of surfactant isolated from BALF was assessed by constrained bubble surfactometry in control and Abca3-cKO mice (E). Data represent mean ± SEM with dot plot overlay. **P ≤ 0.01, ***P ≤ 0.001, and ****P < 0.00001 compared with control as determined by 1-way ANOVA, n = 3–8/group.
Decreased phospholipids and surfactant function after deletion of Abca3.
To determine if depletion of surfactant lipids contributed to the respiratory distress in Abca3-cKO mice, total phospholipid and saturated phosphatidylcholine (SatPC) were measured in BALF. Total phospholipids (Figure 2C) and SatPC (Figure 2D) were significantly decreased after 7 days of tamoxifen (7- and 8-fold reduction, respectively) and were further decreased after 10 days (14- and 25-fold reduction, respectively). Phosphatidylcholine (PC), phosphatidylglycerol (PG), and diacylglycerol (DG) species were decreased (Supplemental Figure 5 and Supplemental Table 1), while phosphatidylinositol (PI) species were increased (Supplemental Figure 5C), consistent with the inverse correlation between PI and PC/PG species previously reported in patients with acute respiratory failure (19, 20). Surfactant function was analyzed by constrained drop surfactometry. Minimum surface tension was unchanged after 6 days; however, minimum surface tension of purified surfactant was increased in Abca3-cKO mice after 10 days (Figure 2E). Normal physiological functions (lung resistance and compliance) were maintained during periods of markedly reduced levels of phospholipids (Supplemental Figure 6); however, loss of surfactant function was consistently associated with respiratory failure 10–15 days after exposure to tamoxifen.
Inflammation and alveolar-capillary leak after deletion of Abca3.
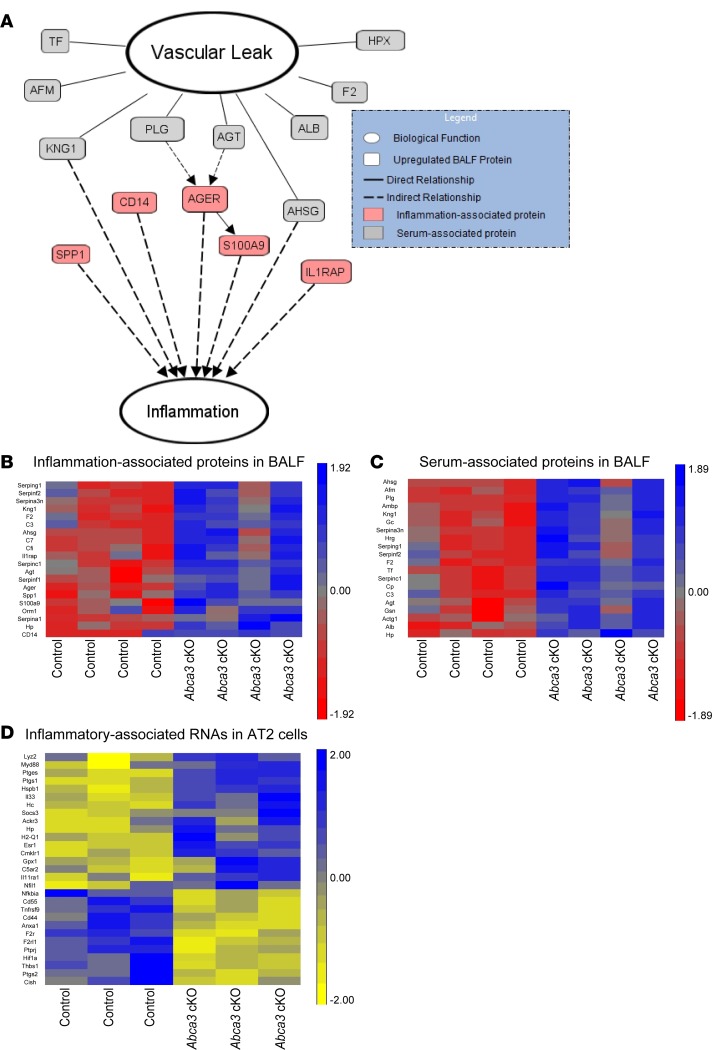
Integrative proteomics and transcriptomics were utilized to identify factors associated with respiratory failure after deletion of Abca3. Epithelial-vascular leak was indicated by increased abundance of serum-associated proteins, including albumin, transferrin, angiotensinogen, coagulation factor F2, and its antagonist plasminogen (PLG) in BALF from Abca3-cKO mice (Figure 3, A–C). Alveolar-capillary leak, with resultant influx of serum proteins, inactivates surfactant (21), likely contributing to respiratory failure after deletion of Abca3. Increased serum proteins in BALF were associated with inflammation and expression of inflammatory mediators, including S100A9 and its receptor AGER (Figure 3, A–C). Proinflammatory signatures in the BALF were complemented by increased Myd88 and Retnla mRNAs (Figure 3D), consistent with the inflammation seen after loss of ABCA3 (Figure 2B). Thus, deletion of Abca3 caused respiratory failure mediated by surfactant deficiency, alveolar-capillary leak, and inflammation. Similarly, deletion of ABCA3-induced RNAs associated with cellular responses to injury and inflammation, including IL-33, Myd88, Socs3, Gpx1, and others in isolated alveolar epithelial cells (Figure 3D).
Figure 3. Inflammation and alveolar-capillary leak after deletion of Abca3.
Integration of proteomic and transcriptomic analyses of control and Abca3-cKO mice. RNAs were measured by RNA-Seq of purified AT2 cells from control and Abca3-cKO mice 6 days after tamoxifen (n = 3). Proteins were analyzed in BALF from control and Abca3-cKO mice 7 days after tamoxifen (n = 4) using liquid chromatography tandem mass spectrometry (LC-MS/MS). (A) Pathway System (GePS) and Ingenuity Pathway Analysis (IPA) suites were used to predict relationships among the significantly increased proteins in BALF; relationships were manually reviewed to ensure relevance before being represented. Heatmaps of inflammation- (B) and serum-associated (C) proteins in BALF and inflammation-associated RNAs in AT2 cells (D) that were differentially expressed in Abca3-cKO mice. Heatmaps were Z-score normalized. Genes and proteins were characterized by ToppGene. RNA changes were > 1.2-fold, P < 0.01. Protein changes were > 1.5-fold, P < 0.05.
Alveolar injury and repair following sublethal deletion of Abca3.
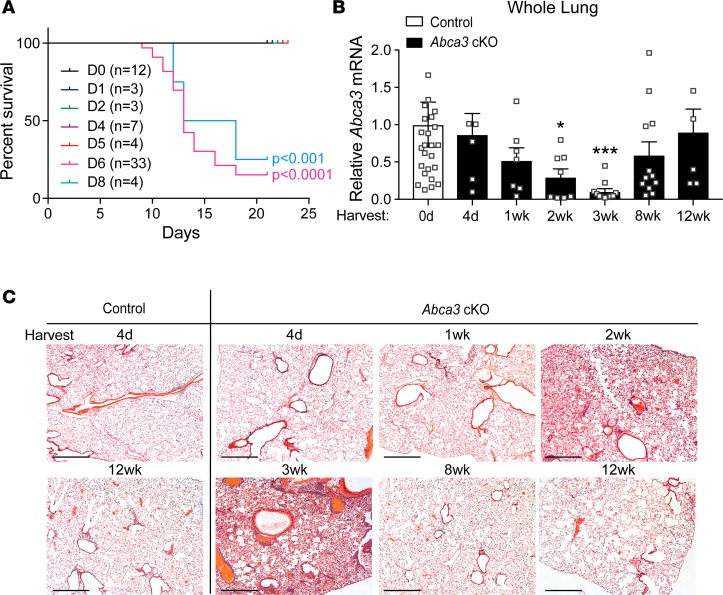
A model of lung repair after deletion of Abca3 was produced by limiting the time of tamoxifen exposure to control mice (Supplemental Figure 7). Administration of tamoxifen for 5 days or less resulted in 100% survival at 21 days (Figure 4A). Abca3 mRNA decreased progressively, being reduced by 90% in whole lung after 3 weeks of recovery (Figure 4B). In spite of extensive deletion, Abca3 mRNA was fully restored at 12 weeks (Figure 4B). Spatially heterogeneous alveolar inflammation and alveolar wall thickening were observed 2–3 weeks after tamoxifen, and lung histopathology, ABCA3 mRNA, and protein staining were fully restored after 12 weeks with the absence of fibrotic remodeling (Figure 4C; Figure 5, A and B; and Supplemental Figure 8), demonstrating selective repopulation of ABCA3-sufficient AT2 cells and remarkable repair of alveolar architecture.
Figure 4. Alveolar injury and regeneration after deletion of Abca3.
(A) Kaplan-Meier curve and statistical analysis (Wilcoxon-Gehan test) for survival of Abca3-cKO mice treated with tamoxifen for 0–8 days. (B) Abca3 mRNA (exon 5–6) expression normalized to β-actin in whole lung from control and Abca3-cKO mice treated with tamoxifen for 4 days with tissue harvest at 4 days to 12 weeks. Data represent Abca3 mRNA with dot plot overlay; n = 5–20 mice/group. *P < 0.05, ***P < 0.0001 compared with control as determined by 1-way ANOVA. (C) Representative lung histology from control and Abca3-cKO mice treated with tamoxifen for 4 days with tissue harvest at 4 days to 12 weeks. Lung tissue from n = 6–8 mice/group were stained by H&E for histological analysis. Scale bars: 500 μm.
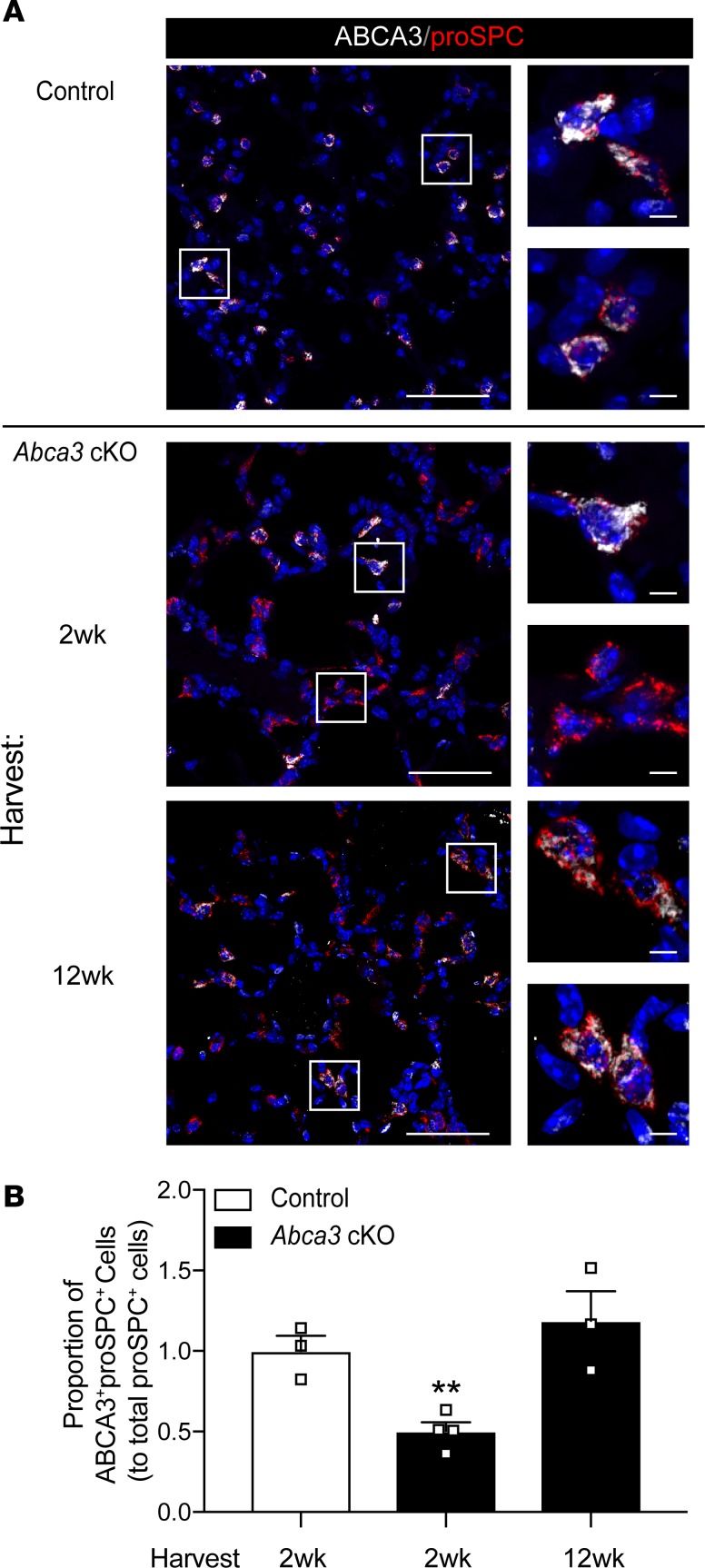
Figure 5. Restoration of ABCA3 expression in AT2 cells after partial deletion.
(A) Representative confocal immunofluorescence for ABCA3 (white) and proSPC (red) in alveolar epithelial cells from control and Abca3-cKO mice treated with tamoxifen for 4 days with tissue harvest at 2 and 12 weeks. Scale bars: 50 μm. Inset cell scale bars: 5 μm. (B) ABCA3+proSPC+ epithelial cells shown in panel A were quantitated using Nikon Elements General Analysis software. Data represent ABCA3+proSPC+ cells, mean ± SEM, n = 3–4 mice/group, **P = 0.0075 as determined by 1-way ANOVA.
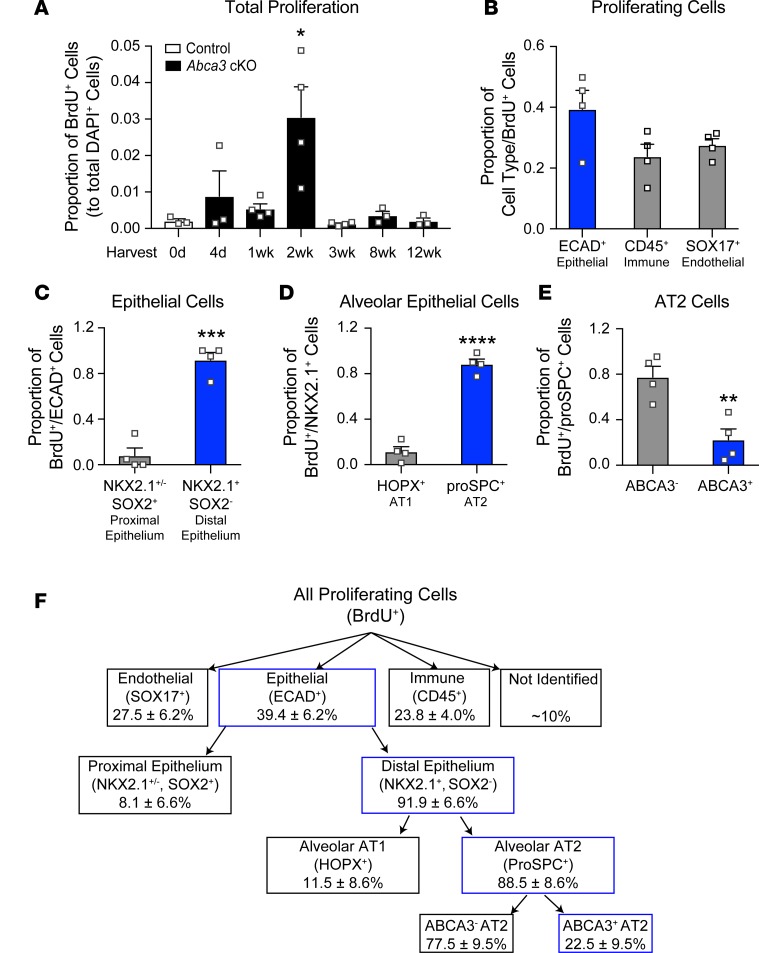
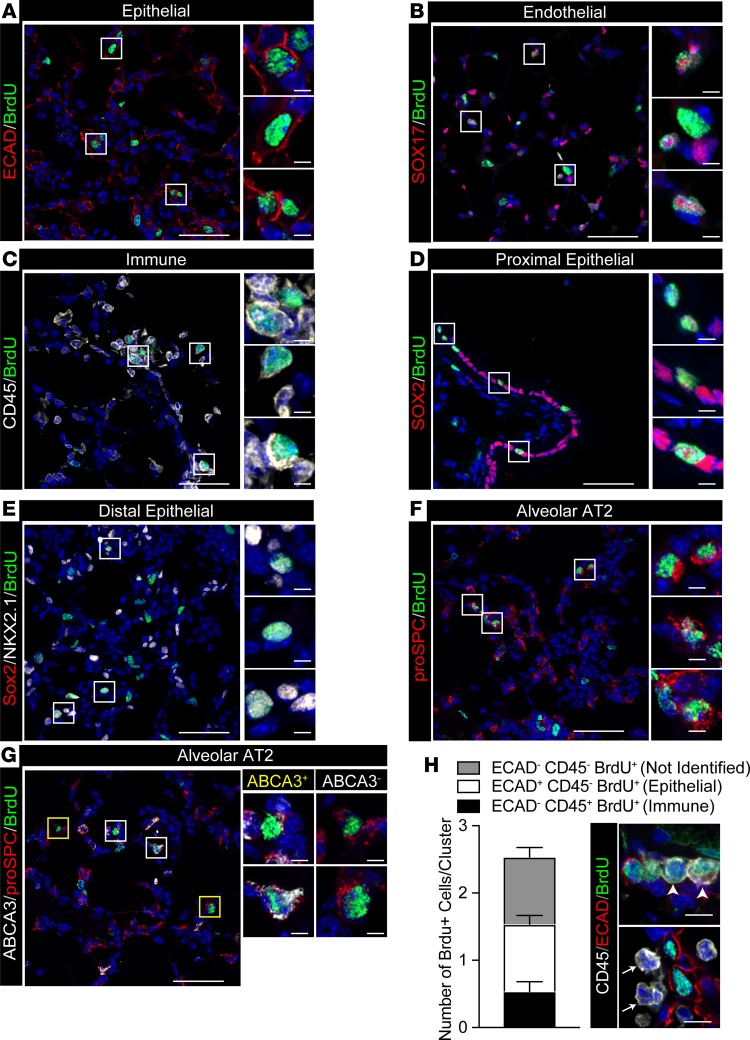
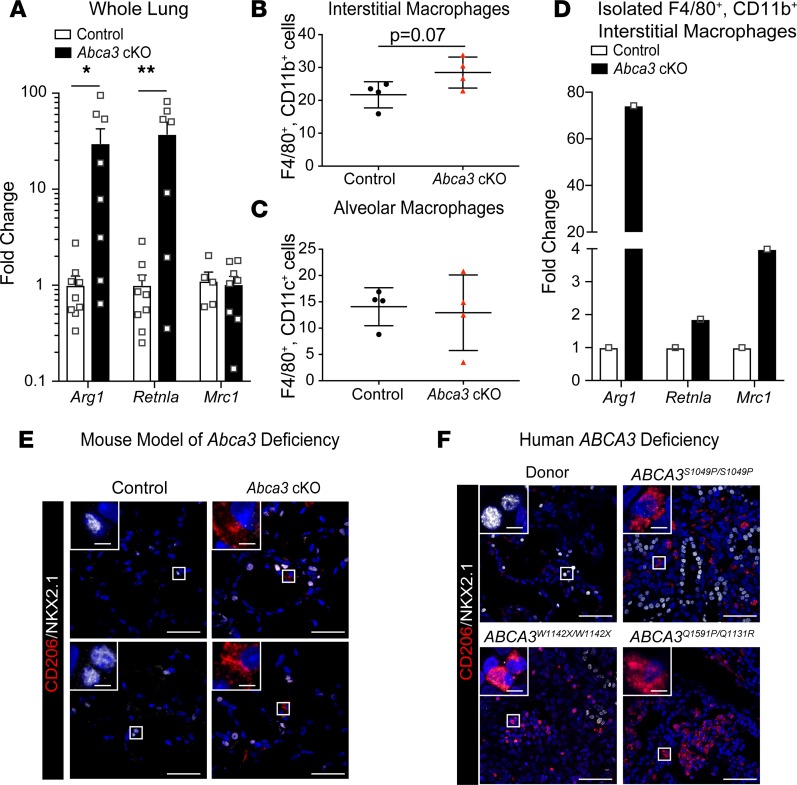
Cell proliferation was quantified following 4 days of tamoxifen (Supplemental Figure 7). BrdU+ cells increased maximally after 2 weeks (Figure 6A), indicating alveolar regeneration. Epithelial (ECAD+) cells accounted for 39.4% ± 6.2% of total BrdU+ cells (Figure 6, B and F, and Figure 7A). Together, endothelial (SOX17+) and immune (CD45+) cells accounted for approximately 50% of BrdU+ cells (Figure 6, B and F, and Figure 7, B and C). Approximately 10% of BrdU+ cells lacked epithelial, endothelial, or BM-derived cell markers (ABCA3–, proSPC–, NKX2.1–, HOPX–, SOX2–, SOX17–, ECAD–, and CD45–), likely indicating stromal cells. The majority of proliferating epithelial cells were present in alveolar regions (NKX2.1+/SOX2–, Figure 6, C and F, and Figure 7, D and E) and were proSPC+ AT2 cells (Figure 6, D and F, and Figure 7F). BrdU-stained SOX2+ cells were confined to conducting airways (Figure 6C and Figure 7D). BrdU-stained HOPX+ cells were rarely detected (Figure 6D), and BrdU-stained KRT5 (basal) cells were not detected in the peripheral lung parenchyma (Supplemental Figure 9). BrdU-stained proSPC+ AT2 cells included both ABCA3+ and ABCA3– cells. Remarkably, ABCA3– cells were the most abundant proliferative AT2 cell type (77.5% ± 9.5%; Figure 6, E and F, and Figure 7G). Taken together, widespread alveolar and bronchial injury caused by deletion of Abca3 was followed by proliferation of diverse cell types that repaired the alveoli. In spite of active proliferation of Abca3-deleted cells, ABCA3-sufficient AT2 cells selectively repopulated the alveoli after 12 weeks, resulting in restoration of ABCA3, lung structure, and function. During active regeneration, proliferating cells were frequently observed in clusters in the damaged alveoli (Figure 7); each BrdU+ cluster generally contained 2–3 BrdU+ cells, with epithelial (ECAD+), immune (CD45+), and stromal cells frequently present within the clusters (Figure 7H). Since atypically activated (M2) macrophages promoted lung regeneration following partial pneumonectomy (22), we assessed markers for M2 macrophages in whole lung 2 weeks following partial loss of Abca3. Arg1 and Retnla were increased 30-fold, while Mrc1 levels were unchanged in whole lung 2 weeks following partial loss of Abca3 (Figure 8A). Arg1 and Retnla expression correlated with increased numbers of CD11b+ (CD45+F4/80+CD11b+CD11c–) but not CD11c+ (CD45+F4/80+CD11c+CD11b–) macrophages (Figure 8, B and C). Furthermore, M2 macrophage markers Arg1, Retnla, and Mrc1 were increased in isolated CD11b+ interstitial macrophages (Figure 8D). Taken together, these data demonstrate that diverse cell types proliferate and interact with lung epithelial cells during alveolar regeneration following loss of Abca3.
Figure 6. Proliferation of alveolar cells following partial deletion of Abca3.
(A) Total cell proliferation was quantitated using Nikon Elements General Analysis software in control and Abca3-cKO mice treated with tamoxifen for 4 days as a proportion of DAPI+ cells. (B) At the peak of proliferation (2 weeks), BrdU+ cells were classified as ECAD+, SOX17+, or CD45+. BrdU+ cells were classified as proximal epithelium (NKX2.1+SOX2+ or NKX2.1–SOX2+) or distal epithelium (NKX2.1+SOX2–) (C), or as AT1 epithelial cells (NKX2.1+HOPX+) or AT2 epithelial cells (NKX2+proSPC+) (D). Proliferating AT2 cells were further defined as ABCA3+ or ABCA3– (E). Data represent mean ± SEM, n = 3–4/group, *P < 0.05 as determined by 1-way ANOVA and **P ≤ 0.01, ***P = 0.0001, and ****P < 0.0001 as determined by 2-tailed Student’s t test. (F) Diagram summarizing proliferating cell types in Abca3-cKO mice. Percentages of proliferating (BrdU+) cells are shown, with data from graphs outlined in blue. Representative images used for quantitation are shown in Figure 7.
Figure 7. Proliferating cells after deletion of Abca3.
Representative confocal immunofluorescence images of proliferating cell clusters analyzed in Figure 6. (A) ECAD+ (red) and BrdU+ (green) epithelial cells, (B) SOX17+ (red) and BrdU+ (green) endothelial cells, (C) CD45+ (white) and BrdU+ (green) immune cells, (D) SOX2+ (red) and BrdU+ (green) proximal epithelial cells, (E) SOX2+ (red), NKX2.1+ (white) and BrdU+ (green) distal epithelial cells, (F) proSPC+ (red) and BrdU+ (green) alveolar AT2 cells, and (G) ABCA3+ (white), proSPC+ (red) and BrdU+ (green) cells. Representative ABCA3+ and ABCA3– AT2 cells are shown in yellow and white boxes, respectively. Scale bars: 50 μm. Inset scale bars: 5 μm. (H) Epithelial (ECAD+) and immune (CD45+) cells were manually counted in BrdU+ clusters. Bars contain ECAD–CD45–BrdU+ (not identified), ECAD+CD45–BrdU+ (epithelial), and ECAD–CD45+BrdU+ (immune) cells superimposed on the total number of proliferating cells per cluster (BrdU+DAPI+). Data are mean ± SEM, n = 3. Representative immunofluorescence of immune cell infiltrates presents in (arrowhead) or bordering (arrow) BrdU+ cell clusters. ECAD (red), CD45 (white), and BrdU (green). Scale bar: 10 μm.
Figure 8. Recruitment of M2-like macrophages in mouse and human ABCA3 deficiency.
(A) Arg1, Retnla, and Mrc1 were measured by reverse transcription PCR (RT-PCR). in whole lung from control and Abca3-cKO mice. Data represent Arg1, Retnla, and Mrc1 mRNA expression with dot plot overlay; n = 7–9 mice/group, *P < 0.05, **P < 0.001 compared with control as determined by 1-way ANOVA. (B and C) Flow cytometry was used to identify interstitial (F4/80+CD11b+) and alveolar (F4/80+CD11c+) macrophages from protease digested lungs of control and Abca3-cKO mice treated with tamoxifen for 4 days harvested at 2 weeks. Data are mean ± SEM, n = 4, unpaired 2-tailed Student’s t test. (D) Arg1, Retnla, and Mrc1 mRNA were measured by RT-PCR in isolated interstitial macrophages (F4/80+CD11b+) from pooled control and Abca3-cKO mice treated with tamoxifen for 4 days harvested at 2 weeks; n = 3 mice/group. (E and F) Representative confocal microscopy of the M2 macrophage marker CD206 (red) and the epithelial marker NKX2.1 (white) in Abca3-cKO mice (n = 3) and pathological tissues from infants with severe lung disease caused by mutations in the ABCA3 gene (n = 5 patients). Scale bars: 50 μm. Inset scale bars: 5 μm.
Recruitment of M2-like macrophages after deletion of Abca3 and in patients with ABCA3-related lung disease.
Pulmonary lesions in ABCA3-related lung disease characteristically contain an abundance of alveolar macrophages located within remodeled alveoli (1, 13). Immunofluorescence confocal microscopy of lung biopsies from patients with severe ABCA3-related lung disease identified inflammatory infiltrates and loss of AT1 cells, with the alveoli and remaining saccules being lined primarily by cuboidal AT2 cells (Figure 8F). Macrophages in these lesions expressed ARG1 and CD206 (Figure 8F and Supplemental Figure 10). Similarly, ARG1+ and CD206+ macrophages were abundant in the regenerating alveoli 2 weeks after partial deletion of Abca3 in the mouse model (Figure 8E and Supplemental Figure 10).
Discussion
While advances have been made in the identification of the genetic causes of interstitial lung diseases in children, development of therapies for these often-lethal disorders is limited by our inadequate knowledge of the pathogenesis of injury and mechanisms of alveolar regeneration. Mutations in ABCA3 are the most prevalent, monogenetic cause of acute and chronic lung disease in infancy (1, 7–9). In the present study, we developed a conditional model of ABCA3 deficiency, enabling analysis of the biochemistry, pulmonary function, and regeneration of the adult mouse lung after the deletion of the Abca3 gene. Loss of ABCA3 in mature AT2 cells caused AT2 cell injury, alveolar-capillary leak, and impaired surfactant homeostasis, resulting in diffuse alveolar damage and subsequent respiratory failure. Extensive but partial deletion of ABCA3 caused severe lung injury, followed by restoration of ABCA3 expression and alveolar structure by residual, nontargeted ABCA3-sufficient cells, resulting in complete restoration of pulmonary homeostasis after 12 weeks. The remarkable selective advantage of the ABCA3-sufficient cells to restore alveolar homeostasis provides the foundation for genetic replacement or correction of ABCA3 deficiency in the future. Likewise, understanding the pathogenesis of the disorder may enable development of therapeutic strategies to minimize alveolitis and maximize regenerative capacity in infants affected by ABCA3 mutations.
Surfactant deficiency and respiratory failure after loss of ABCA3.
The biochemical response to deletion of ABCA3 in the adult lung is consistent with the known role of ABCA3 in surfactant homeostasis in perinatal adaptation, with respiratory failure being associated with decreased alveolar phospholipids following birth (1, 9, 13). ABCA3 is located in limiting membranes of the lamellar bodies, where it mediates transport of phospholipids, primarily PC and PG, into the organelle (5, 23–25). Consistent with previous findings in patients with ABCA3 deficiency (19, 20), alveolar PC and PG decreased after deletion of ABCA3. Remarkably, surfactant function and lung function were maintained for a prolonged period, during which surfactant lipids were markedly decreased. Proteomic analyses of BALF, before respiratory distress, demonstrated increased concentrations of serum proteins, indicating alveolar-capillary leak. Paucity of surfactant and the influx of plasma proteins known to inactivate surfactant activity (21) likely explain the respiratory failure that followed.
Inflammatory responses after ABCA3 gene deletion.
Deletion of ABCA3 in AT2 cells caused inflammation, alveolar capillary leak, and decreased surfactant phospholipid concentrations prior to respiratory failure. A diversity of serum and parenchymal proteins were increased in BALF. Serum proteins in the alveoli that inhibit surfactant activity (21), together with the paucity of phospholipids, creates mechanical stress that causes parenchymal damage. Many of the serum proteins identified in BALF recruit and activate inflammatory cells (26, 27). Thus, both mechanical forces and inflammation likely contribute to the alveolitis and ensuing respiratory failure seen after the loss of ABCA3. AGER, S100A9, CD14, IL1RAP, SPP1, and SerpinA1 mediate macrophage or neutrophil recruitment and activation, whether derived from inflammatory or parenchymal cells (28–32). Serum proteins that activate macrophages were present in the BALF, including albumin, AGT, SerpinF1, GSN, GC, HRG, and KNG1 (27, 33–35). RNA analysis of isolated alveolar epithelial cells from the Abca3-deleted mice indicated increased expression of RNAs associated with cell injury and induced inflammatory response, including IL-33. Taken together, intrinsic AT2 cell dysfunction, tissue injury (stemming from the loss of surfactant activity, changes in mechanical forces in the alveolus, and alveolar capillary leak), and the presence of serum proteins that both inhibit surfactant function and activate inflammation likely contribute to respiratory failure after loss of ABCA3.
Alveolar repair after partial deletion of ABCA3.
We utilized Sftpc-CreERT2 to selectively delete Abca3 in AT2 cells, resulting in a virtually complete loss of ABCA3 mRNA and protein within 6–7 days of tamoxifen, followed by respiratory failure. Following partial loss of Abca3, severe lung injury and inflammation were accompanied by rapid proliferation of multiple cell types, including endothelial (SOX17+), epithelial (ECAD+), inflammatory (CD45+), alveolar (proSPC+ and ABCA3+), and conducting airway epithelial (SOX2+) cells. In spite of active proliferation of both Abca3-sufficient and -deficient cells, following the regenerative phase, ABCA3-expressing cells were more abundant, suggesting a selective advantage of ABCA3-expressing AT2 cells during lung regeneration. Since Cre-mediated recombination is not reversible, it is most likely that the ABCA3-expressing cells were derived from AT2 cells that had not undergone Cre-mediated recombination. Alternatively, these cells may be derived from other progenitors that did not express the Sftpc-CreERT2. Taken together, these data support the feasibility of restoring functional ABCA3 in AT2 cells, where survival and proliferation would enable development of future therapies for lung disease. Currently, lung transplantation is the only effective treatment for severe lung disease caused by mutations in ABCA3, a challenging process for infants with chronic respiratory failure (12, 13). BrdU labeling was observed in SOX2+ cells; these cells were confined to conducting airways. KRT5+ basal cells, known to proliferate following severe influenza infection (36, 37), were not observed in either alveolar or conducting airways. BrdU labeling was more numerous in ABCA3-deficient AT2 cells, perhaps indicating increased cell turnover; however, restoration of ABCA3 expression at 12 weeks indicated a selective advantage of the ABCA3-sufficient cells.
During repair, BrdU incorporation was present in inflammatory, endothelial, epithelial, and stromal cells, and it was consistently observed in cell clusters containing diverse cell types. Interactions between alveolar macrophages and AT2 cells were previously suggested to enhance cell survival, via mitochondrial transfer or suppression of inflammation (22, 38–40). In the present study, CD45+ macrophages were frequently found in close proximity to proliferating AT2 cells. Cytometric analysis demonstrated increased numbers of CD11b+ macrophages and expression of RNAs associated with M2 macrophage activation during regeneration. Studies of lung regeneration after partial pneumonectomy support the concept that interactions among activated macrophages and lung parenchymal cells create a proliferative microenvironment (22). In the present study, regeneration was associated with marked increase in inflammatory cell expression of ARG1 and other markers of atypical macrophage activation (M2-like), including RETNLA and CD206, proposed to contribute to repair processes (22, 41, 42). Alveolar macrophages expressing CD206 and ARG1 were identified by immunofluorescence confocal microscopy in both Abca3-cKO mouse lung during regeneration and in lung tissue from patients with severe lung disease caused by ABCA3 mutations. While M2 monocytic–derived macrophages have been implicated in profibrotic processes (43), we did not observe fibrotic remodeling in the present mouse model. Recruitment of macrophages is a common feature of patients with interstitial lung disease (13, 44) and in mouse models of surfactant deficiency caused by disruption of surfactant lipid or protein homeostasis. For example, chronic AT2 cell injury caused by inhibition of SREBP, a transcriptional regulator of lipid homeostasis in AT2 cells, was associated with inflammation, expression of ARG1, and recruitment of atypical macrophages (45). Whether the recruitment of M2 macrophages to sites of alveolar cell proliferation contributes to repair or injury in the context of ABCA3-mediated lung disease will require further investigation.
In summary, the present study highlights the remarkable regenerative capacity of ABCA3-sufficient AT2 cells, a selective advantage that supports strategies to replace, repair, or improve the function of ABCA3 for treatment of respiratory failure caused by ABCA3 mutations. Likewise, knowledge regarding the pathogenesis of respiratory failure, alveolar-capillary leak, and inflammation may provide opportunities to minimize lung injury caused by disorders of surfactant homeostasis after alveolar injury.
Methods
Mouse model.
Homozygous Abca3-floxed (Abca3flox/flox) mice were produced in our laboratory as previously described (17). In Abca3flox/flox mice, LoxP sites flank exons 4 and 7 of the Abca3 gene. These exons encode transmembrane domain 1 and the first extramembranous loop of ABCA3. Abca3flox/flox mice were mated to the Sftpc-CreERT2 mice (gift from B. Hogan, Duke University, Durham, North Carolina, USA) (46) that express a tamoxifen-inducible Cre (IRES-CreERT2) under the control of the Sftpc promoter–generating control mice. Genotyping was determined through allele-specific PCR from tail biopsies using Abca3- and Sftpc-CreERT2–specific primers (Supplemental Table 2). To activate Cre recombination, control age-matched adult (6- to 8-week-old) mice were administered a tamoxifen diet (400 mg tamoxifen citrate/kg, ENVIGO) ad libitum. In the presence of tamoxifen, exons 4–7 of the Abca3 gene were deleted from AT2 cells of control mice, termed Abca3-cKO mice. Mice with signs of respiratory distress as assessed by dehydration, dyspnea, or hunched appearance accompanied by lethargy were sacrificed in accordance with protocols approved by the IACUC of the Cincinnati Children’s Hospital Research Foundation (Cincinnati, Ohio, USA). For lineage-tracing studies, Sftpc-CreERT2 mice were mated to Gt(ROSA)26SortdTomato (The Jackson Laboratory), generating Sftpc-CreERT2;tdTomato mice (47). For partial deletion studies, control (untreated) and Abca3-cKO mice were administered a tamoxifen diet for 1–8 days and then returned to normal chow until tissue harvest (Supplemental Figure 7). For proliferation studies, control and Abca3-cKO mice were administered 50 μg/g body weight BrdU (BD Biosciences; 550891) i.p. 2 hours prior to tissue harvest (Supplemental Figure 7). All mice were maintained in a pathogen-free environment.
Histology and immunofluorescence.
Lungs were inflation fixed at 25 cm H2O in 4% paraformaldehyde, embedded in paraffin or OCT compound, and sectioned (5–7 μm). Paraffin tissue sections were stained with H&E according to standard protocols. For immunofluorescence, primary antibodies are listed in Supplemental Table 2. Rabbit ABCA3 antibody (Seven Hills Bioreagents; Supplemental Table 2) was used in Figure 1; however, the guinea pig ABCA3 antibody (in-house; Supplemental Table 2) was used in the remainder of the paper due to antibody compatibility with costains. Secondary antibodies were used at 1:200 dilutions; those antibodies include donkey anti–mouse IgG 488 (A21202), donkey anti–rabbit IgG 488 (A21206), donkey anti–rabbit IgG 647 (A31573), donkey anti–goat IgG 568 (A11057), donkey anti–goat IgG 647 (A21447), and donkey anti–rabbit 568 (A11011) (all from Invitrogen); donkey anti–rat IgG 568 (ab175475, Abcam); and donkey anti–guinea pig IgG 647 (706-605-148, Jackson ImmunoResearch). In all images, background was reduced and colors were leveled in Photoshop Elements 14.
Human lung tissue.
Excess lung tissue was obtained from deidentified normal adult tissue donors (provided by Scott Randell, University of North Carolina, Chapel Hill, North Carolina, USA), lung biopsies of deidentified infants with ABCA3 gene mutations (provided by A. Francine Tryka, Arkansas Children’s Hospital, Little Rock, Arizona, USA; and L. Nogee, Johns Hopkins Children’s Center, Baltimore, Maryland, USA) (48), and lung explants (Division of Pathology, Cincinnati Children’s Hospital Medical Center) under protocols approved by the IRBs of each institution for the use of human tissue samples (Supplemental Table 3).
Morphometric analyses.
Heterogeneous cell populations from frozen immunofluorescent sections were characterized in Nikon Elements General Analysis 4.50. Widefield tile scans were taken of the left lobe on a Nikon Eclipse Ni-E upright microscope, 10–15 20× random alveolar regions were selected. In each image, an average of 1,000 cells were counted for analysis. A total of 10,000–15,000 cells were counted per mouse. To determine the composition of proliferating cell clusters, 20–40 proliferating (BrdU+) clusters were manually counted.
Phenotypic markers were split over the 3 immunofluorescent channels (FITC, Texas-Red, Cy5). For each image, individual cellular regions (nuclear, cytoplasmic, and membrane) were extracted to determine differences in subpopulations normalized to nuclear DAPI staining. Nuclear DAPI+ cells (total cells) were selected using bright spot detection (diameter, 6.583; contrast, 140 for a 20× image). To eliminate false negatives, the nucleus was grown to encompass a larger cytoplasmic area. Each of these cellular regions was extracted by setting an intensity threshold for cytoplasmic and nuclear stains (membrane stains were extracted with bright spot detection). Individual phenotypic markers were then analyzed according to colocalization/overlap with DAPI+ cells. To eliminate error due to cytoplasmic or membrane cellular regions overlapping multiple nuclei, every cell positive for a phenotypic marker was filtered by assessing the area of overlap with DAPI+ and intensity of cellular region. Once subpopulations of each single phenotypic marker was created, every double/triple stain was analyzed by comparisons of area of colocalization/overlap. Counts were verified manually. No statistical difference was found between Nikon Elements counts and manual counts (r2 > 0.9). Subpopulations were separated by cell type and region. A 2-tailed Student’s t test was run to determine statistical differences between related phenotypes.
AT2 cell purification.
AT2 cells were purified by cell depletion as described by Corti et al. (49). Lungs were perfused with 0.9% saline through the pulmonary artery, tracheostomized with a 20-gauge luer stub, instilled with 2 ml Dispase (Corning) resected, and incubated at 37°C for 6 minutes. Lung lobes were separated and placed in gentleMACS C tubes (Miltenyi Biotec Inc.) with DMEM and 100 U/ml DNAse to generate a single cell suspension with a MACS Dissociator. Cell suspensions were centrifuged at 430 g for 6 minutes, washed with 10 ml DMEM media, and filtered through 40-μm cell strainers. The filtered cell suspensions were centrifuged at 430 g for 6 minutes, and resuspended in 500 μl MACs + 0.5% BSA buffer (Miltenyi Biotec Inc.) with biotinylated anti-mouse antibodies as listed in Supplemental Table 2 for 30 minutes. Cell suspensions were washed with MACs + 0.5% BSA buffer, centrifuged at 430 g, resuspended in 180 μl MACs + 0.5% BSA buffer containing 20 μl anti-biotin microbeads (Miltenyi Biotec Inc.), and purified over a LS column (Miltenyi Biotec Inc.). Purified AT2 cells (CD45–, CD16/32–, Ter119–, CD90–, and CD31–) were resuspended in RLT Buffer (RNeasy Plus Mini Kit, Qiagen) and stored at –80°C.
BALF.
BALF was collected by intratracheal intubation and 5 serial lung lavages using 1 ml 0.9% saline + 0.5 mM EDTA; combined lung lavage was placed on ice. Inflammatory cells were isolated from the BALF by centrifugation at 280 g for 10 minutes, and resuspended in 100 μl 0.9% saline for total and differential cell analyses. Cell-free BALF (supernatant) was used for either SatPC analysis or surfactant function measurements following isolation of surfactant.
RNA isolation.
Total RNA was isolated from whole lung or isolated adult AT2 cells with the RNeasy Plus Mini Kit (Qiagen). Total RNA was isolated from sorted CD11b+F4/80+ alveolar macrophages with the Single Cell RNA Purification Kit (Norgen Biotech Corp.).
Real-time PCR.
RNA (500 ng) was reverse transcribed into cDNA using the iScript kit (Bio-Rad). Quantitative PCR reactions were performed in triplicate using TaqMan gene expression master mix (Applied Biosystems) on a StepOnePlus real-time PCR system (Applied Biosystems). Expression was normalized to endogenous ACTB or 18S; probe sets are listed in Supplemental Table 2.
Transcriptomic, proteomic, and lipidomic analyses.
RNA sequencing (RNA-Seq) was performed on purified AT2 cells (CD45–, CD16/32–, Ter119–, CD90–, and CD31–) from control and Abca3-cKO mice 6 days after tamoxifen. RNA-Seq was performed utilizing the Ovation RNA-Seq System v2 (NuGEN) and Nextera XT DNA Sample Preparation kits (Illumina Technologies) by the Cincinnati Children’s Hospital Medical Center’s Gene Expression Core. RNA-Seq FASTQ files were aligned to the mouse genome version mm10 using the Bioconductor’s Genomic Alignment software (50). Differential gene expression was evaluated using DeSeq software (51). Genes were deemed differentially expressed with a nbinom test P < 0.05, and fold change cut-off of > 1.2 with reads per kilobase million (RPKM) > 1 for all samples in at least 1 condition (50, 51).
Lipidomic analysis was performed on BALF at PNNL as previously described (52). Proteomics analysis was performed on BALF at Pacific Northwest Laboratory using a label-free approach (53) as detailed in Supplemental Methods. Two-tailed Student’s t tests were used to determine significantly changed lipid species and proteins. All significantly changed proteins and lipids had a P < 0.01 and a fold change > 1.2. RNAs, proteins, and lipids were further analyzed using functional genomics tools. ToppGene was used to identify functional enrichments of significantly altered RNAs and proteins (54). Genomatix Pathway System (GePS) and Ingenuity Pathway Analysis (IPA) suites were used to predict relationships among differentially expressed genes and proteins based on literature mining (www.genomatix.de and www.qiagen.com/ingenuity). Predicted relationships were then manually reviewed. Partek Genomic Suites (Partek Incorporated) were used to generate heatmaps representing significant changes. System models were created using IPA’s Path Designer.
Total and differential cell counts.
Cytospin slides of ~1 × 106 BALF cells were stained with the Siemens Diff-Quik stain kit (ThermoFisher Scientific, NC0674866). A total of 300–400 cells were counted per slide and classified as monocyte/macrophage, lymphocyte, or neutrophil.
SatPC.
Lipids were extracted from the cell-free BALF with chloroform and methanol (55). SatPC was isolated using the osmium tetroxide-based method described by Mason et al. (56).
Surfactant function.
Surfactant was isolated from the cell-free BALF by ultracentrifugation at 40,000 g for 20 minutes at 4°C. Pelleted surfactant was resuspended in 100 μl 0.9% saline, aliquoted, and stored at –20°C. Surfactant function was assessed by Constrained Sessile Drop Surfactometry on 9–10 μl drops of 2 mg/ml phospholipid cycled at 30 cycles/minute to determine minimal surface tension (57). Measurements reflect data from individual mice for control and Abca3-cKO mice, except for day-10 tamoxifen–treated samples. Samples from this group were pooled, due to low surfactant levels, to obtain enough material for analysis.
FACS analysis.
Cytometric measurements of M2 macrophages were conducted on single cell suspensions of whole lungs as previously described (22). Following perfusion, lungs were inflated with 2 ml protease solution containing collagenase type II (Worthington Biochemical Corporation, at 450 U/ml), elastase (Worthington Biochemical Corporation, at 4U/ml), and dispase (Corning, 50 U/ml). Lungs were resected and incubated at room temperature for 45 minutes. Lung lobes were separated and teased apart in the presence of 100 U/ml DNAse (MilliporeSigma), and single cell suspensions were generated by gentle pipetting. Cell suspensions were filtered through 40 μm cell strainers. Cells were incubated with 1 ml RBC lysis buffer (MilliporeSigma) at room temperature for 2 minutes and washed with FACS buffer (PBS + 2% FBS). Cells were kept on ice and stained. Cells were blocked with CD16/CD32 Fc receptor block (eBiosciences, catalog 14-0161-85, clone 93) at 1:100 for 10 minutes and washed. Cells were incubated for 30 minutes with 1:50 fluorophore-conjugated antibodies as listed in Supplemental Table 2. Flow cytometry was performed on a BD LSR Fortessa, and sorting was performed on a BD FACS Aria II. M2-alveolar macrophages (Cd11b+F4/80+) were isolated and stored in RNeasy Lysis Buffer (RLT Plus, Qiagen) for RNA analysis. Compensation was performed using single-channel controls using OneComp eBeads (ThermoFisher Scientific). Gating controls for each channel were determined by FMOs (fluorescence minus one). Data were analyzed with FlowJo 10.0.8 software.
Statistics.
Data are mean ± SEM, with 1-way ANOVA or 2-tailed Student’s t test. Statistical analyses were performed in GraphPad Prism 7 (GraphPad Software, Inc.).
Study approval.
Mice were housed in pathogen-free facilities according to protocols approved by the IACUC of the Cincinnati Children’s Hospital Research Foundation. Human lung tissue/lung biopsies were provided by Scott Randell (University of North Carolina, Chapel Hill, North Carolina, USA), A. Francine Tryka (Arkansas Children’s Hospital), L. Nogee (Johns Hopkins Children’s Center), and the Division of Pathology (Cincinnati Children’s Hospital Medical Center) under protocols approved by the IRBs of each institution.
Author contributions
JPB and JAW developed concepts; JPB, JAW, and TNR developed experiments, interpreted data, and prepared the manuscript; TNR, CAS, ALF, KMB, WZ, RV, EMZ, SED, GC, and CA conducted experiments; and JMS and YX performed bioinformatics analyses.
Supplementary Material
Acknowledgments
This work was supported by NIH grants U0110964 and U0122642 (JAW). Portions of this work were supported by U01-HL-122703 and performed in W. R. Wiley Environmental Molecular Sciences Laboratory (EMSL), a Department of Energy (DOE) office of Biological and Environmental Research (BER) national user facility located at PNNL. The RNA-Seq data are available at the Gene Expression Omnibus (GEO) database, accession number GSE103506. The lipidomics and proteomics data are available at the MassIVE data repository, MSV000081473.
Version 1. 12/21/2017
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2017;2(24):e97381. https://doi.org/10.1172/jci.insight.97381.
Contributor Information
Tara N. Rindler, Email: tara.rindler@cchmc.org.
John M. Snowball, Email: john.snowball@cchmc.org.
Wenjia Zhou, Email: wzhou67@uwo.ca.
Ruud Veldhuizen, Email: rveldhui@uwo.ca.
Geremy Clair, Email: geremy.clair@pnnl.gov.
Yan Xu, Email: yan.xu@cchmc.org.
References
- 1.Whitsett JA, Wert SE, Weaver TE. Diseases of pulmonary surfactant homeostasis. Annu Rev Pathol. 2015;10:371–393. doi: 10.1146/annurev-pathol-012513-104644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wambach JA, et al. Genotype-phenotype correlations for infants and children with ABCA3 deficiency. Am J Respir Crit Care Med. 2014;189(12):1538–1543. doi: 10.1164/rccm.201402-0342OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamano G, et al. ABCA3 is a lamellar body membrane protein in human lung alveolar type II cells. FEBS Lett. 2001;508(2):221–225. doi: 10.1016/S0014-5793(01)03056-3. [DOI] [PubMed] [Google Scholar]
- 4.Klugbauer N, Hofmann F. Primary structure of a novel ABC transporter with a chromosomal localization on the band encoding the multidrug resistance-associated protein. FEBS Lett. 1996;391(1-2):61–65. doi: 10.1016/0014-5793(96)00700-4. [DOI] [PubMed] [Google Scholar]
- 5.Cheong N, et al. ABCA3 is critical for lamellar body biogenesis in vivo. J Biol Chem. 2007;282(33):23811–23817. doi: 10.1074/jbc.M703927200. [DOI] [PubMed] [Google Scholar]
- 6.Ban N, et al. ABCA3 as a lipid transporter in pulmonary surfactant biogenesis. J Biol Chem. 2007;282(13):9628–9634. doi: 10.1074/jbc.M611767200. [DOI] [PubMed] [Google Scholar]
- 7.Shulenin S, Nogee LM, Annilo T, Wert SE, Whitsett JA, Dean M. ABCA3 gene mutations in newborns with fatal surfactant deficiency. N Engl J Med. 2004;350(13):1296–1303. doi: 10.1056/NEJMoa032178. [DOI] [PubMed] [Google Scholar]
- 8.Bullard JE, Wert SE, Whitsett JA, Dean M, Nogee LM. ABCA3 mutations associated with pediatric interstitial lung disease. Am J Respir Crit Care Med. 2005;172(8):1026–1031. doi: 10.1164/rccm.200503-504OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kröner C, et al. Lung disease caused by ABCA3 mutations. Thorax. 2017;72(3):213–220. doi: 10.1136/thoraxjnl-2016-208649. [DOI] [PubMed] [Google Scholar]
- 10.Wambach JA, et al. Functional Characterization of ATP-Binding Cassette Transporter A3 Mutations from Infants with Respiratory Distress Syndrome. Am J Respir Cell Mol Biol. 2016;55(5):716–721. doi: 10.1165/rcmb.2016-0008OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brasch F, et al. Alteration of the pulmonary surfactant system in full-term infants with hereditary ABCA3 deficiency. Am J Respir Crit Care Med. 2006;174(5):571–580. doi: 10.1164/rccm.200509-1535OC. [DOI] [PubMed] [Google Scholar]
- 12.Eldridge WB, et al. Outcomes of Lung Transplantation for Infants and Children with Genetic Disorders of Surfactant Metabolism. J Pediatr. 2017;184:157–164.e2. doi: 10.1016/j.jpeds.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doan ML, et al. Clinical, radiological and pathological features of ABCA3 mutations in children. Thorax. 2008;63(4):366–373. doi: 10.1136/thx.2007.083766. [DOI] [PubMed] [Google Scholar]
- 14.Ochs M, et al. The number of alveoli in the human lung. Am J Respir Crit Care Med. 2004;169(1):120–124. doi: 10.1164/rccm.200308-1107OC. [DOI] [PubMed] [Google Scholar]
- 15.Castranova V, Rabovsky J, Tucker JH, Miles PR. The alveolar type II epithelial cell: a multifunctional pneumocyte. Toxicol Appl Pharmacol. 1988;93(3):472–483. doi: 10.1016/0041-008X(88)90051-8. [DOI] [PubMed] [Google Scholar]
- 16.Hogan BL, et al. Repair and regeneration of the respiratory system: complexity, plasticity, and mechanisms of lung stem cell function. Cell Stem Cell. 2014;15(2):123–138. doi: 10.1016/j.stem.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Besnard V, et al. Conditional deletion of Abca3 in alveolar type II cells alters surfactant homeostasis in newborn and adult mice. Am J Physiol Lung Cell Mol Physiol. 2010;298(5):L646–L659. doi: 10.1152/ajplung.00409.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barkauskas CE, et al. Type 2 alveolar cells are stem cells in adult lung. J Clin Invest. 2013;123(7):3025–3036. doi: 10.1172/JCI68782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Günther A, et al. Surfactant alterations in severe pneumonia, acute respiratory distress syndrome, and cardiogenic lung edema. Am J Respir Crit Care Med. 1996;153(1):176–184. doi: 10.1164/ajrccm.153.1.8542113. [DOI] [PubMed] [Google Scholar]
- 20.Pison U, et al. Surfactant abnormalities in patients with respiratory failure after multiple trauma. Am Rev Respir Dis. 1989;140(4):1033–1039. doi: 10.1164/ajrccm/140.4.1033. [DOI] [PubMed] [Google Scholar]
- 21.Berry D, Ikegami M, Jobe A. Respiratory distress and surfactant inhibition following vagotomy in rabbits. J Appl Physiol. 1986;61(5):1741–1748. doi: 10.1152/jappl.1986.61.5.1741. [DOI] [PubMed] [Google Scholar]
- 22.Lechner AJ, et al. Recruited Monocytes and Type 2 Immunity Promote Lung Regeneration following Pneumonectomy. Cell Stem Cell. 2017;21(1):120–134.e7. doi: 10.1016/j.stem.2017.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mulugeta S, et al. Identification of LBM180, a lamellar body limiting membrane protein of alveolar type II cells, as the ABC transporter protein ABCA3. J Biol Chem. 2002;277(25):22147–22155. doi: 10.1074/jbc.M201812200. [DOI] [PubMed] [Google Scholar]
- 24.Fitzgerald ML, et al. ABCA3 inactivation in mice causes respiratory failure, loss of pulmonary surfactant, and depletion of lung phosphatidylglycerol. J Lipid Res. 2007;48(3):621–632. doi: 10.1194/jlr.M600449-JLR200. [DOI] [PubMed] [Google Scholar]
- 25.Hammel M, et al. Targeted inactivation of the murine Abca3 gene leads to respiratory failure in newborns with defective lamellar bodies. Biochem Biophys Res Commun. 2007;359(4):947–951. doi: 10.1016/j.bbrc.2007.05.219. [DOI] [PubMed] [Google Scholar]
- 26.Christofidou-Solomidou M, et al. Recombinant plasma gelsolin diminishes the acute inflammatory response to hyperoxia in mice. J Investig Med. 2002;50(1):54–60. doi: 10.2310/6650.2002.33518. [DOI] [PubMed] [Google Scholar]
- 27.Yang Z, Chiou TT, Stossel TP, Kobzik L. Plasma gelsolin improves lung host defense against pneumonia by enhancing macrophage NOS3 function. Am J Physiol Lung Cell Mol Physiol. 2015;309(1):L11–L16. doi: 10.1152/ajplung.00094.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van den Brule S, et al. Lung inflammation and thymic atrophy after bleomycin are controlled by the prostaglandin D2 receptor DP1. Am J Respir Cell Mol Biol. 2014;50(1):212–222. doi: 10.1165/rcmb.2012-0520OC. [DOI] [PubMed] [Google Scholar]
- 29.Chavakis T, et al. The pattern recognition receptor (RAGE) is a counterreceptor for leukocyte integrins: a novel pathway for inflammatory cell recruitment. J Exp Med. 2003;198(10):1507–1515. doi: 10.1084/jem.20030800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ryckman C, Vandal K, Rouleau P, Talbot M, Tessier PA. Proinflammatory activities of S100: proteins S100A8, S100A9, and S100A8/A9 induce neutrophil chemotaxis and adhesion. J Immunol. 2003;170(6):3233–3242. doi: 10.4049/jimmunol.170.6.3233. [DOI] [PubMed] [Google Scholar]
- 31.Jeyaseelan S, Chu HW, Young SK, Freeman MW, Worthen GS. Distinct roles of pattern recognition receptors CD14 and Toll-like receptor 4 in acute lung injury. Infect Immun. 2005;73(3):1754–1763. doi: 10.1128/IAI.73.3.1754-1763.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahadeva R, et al. Polymers of Z alpha1-antitrypsin co-localize with neutrophils in emphysematous alveoli and are chemotactic in vivo. Am J Pathol. 2005;166(2):377–386. doi: 10.1016/S0002-9440(10)62261-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cortez-Retamozo V, et al. Angiotensin II drives the production of tumor-promoting macrophages. Immunity. 2013;38(2):296–308. doi: 10.1016/j.immuni.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chavan SS, et al. Identification of pigment epithelium-derived factor as an adipocyte-derived inflammatory factor. Mol Med. 2012;18:1161–1168. doi: 10.2119/molmed.2012.00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamamoto N, Homma S. Vitamin D3 binding protein (group-specific component) is a precursor for the macrophage-activating signal factor from lysophosphatidylcholine-treated lymphocytes. Proc Natl Acad Sci USA. 1991;88(19):8539–8543. doi: 10.1073/pnas.88.19.8539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zuo W, et al. p63(+)Krt5(+) distal airway stem cells are essential for lung regeneration. Nature. 2015;517(7536):616–620. doi: 10.1038/nature13903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vaughan AE, et al. Lineage-negative progenitors mobilize to regenerate lung epithelium after major injury. Nature. 2015;517(7536):621–625. doi: 10.1038/nature14112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Islam MN, et al. Mitochondrial transfer from bone-marrow-derived stromal cells to pulmonary alveoli protects against acute lung injury. Nat Med. 2012;18(5):759–765. doi: 10.1038/nm.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jackson MV, et al. Mitochondrial Transfer via Tunneling Nanotubes is an Important Mechanism by Which Mesenchymal Stem Cells Enhance Macrophage Phagocytosis in the In Vitro and In Vivo Models of ARDS. Stem Cells. 2016;34(8):2210–2223. doi: 10.1002/stem.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bhattacharya J, Westphalen K. Macrophage-epithelial interactions in pulmonary alveoli. Semin Immunopathol. 2016;38(4):461–469. doi: 10.1007/s00281-016-0569-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Monticelli LA, et al. Arginase 1 is an innate lymphoid-cell-intrinsic metabolic checkpoint controlling type 2 inflammation. Nat Immunol. 2016;17(6):656–665. doi: 10.1038/ni.3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aggarwal NR, King LS, D’Alessio FR. Diverse macrophage populations mediate acute lung inflammation and resolution. Am J Physiol Lung Cell Mol Physiol. 2014;306(8):L709–L725. doi: 10.1152/ajplung.00341.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Misharin AV, et al. Monocyte-derived alveolar macrophages drive lung fibrosis and persist in the lung over the life span. J Exp Med. 2017;214(8):2387–2404. doi: 10.1084/jem.20162152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wert SE, Whitsett JA, Nogee LM. Genetic disorders of surfactant dysfunction. Pediatr Dev Pathol. 2009;12(4):253–274. doi: 10.2350/09-01-0586.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Plantier L, et al. Activation of sterol-response element-binding proteins (SREBP) in alveolar type II cells enhances lipogenesis causing pulmonary lipotoxicity. J Biol Chem. 2012;287(13):10099–10114. doi: 10.1074/jbc.M111.303669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu X, et al. Evidence for type II cells as cells of origin of K-Ras-induced distal lung adenocarcinoma. Proc Natl Acad Sci USA. 2012;109(13):4910–4915. doi: 10.1073/pnas.1112499109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13(1):133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tryka AF, Wert SE, Mazursky JE, Arrington RW, Nogee LM. Absence of lamellar bodies with accumulation of dense bodies characterizes a novel form of congenital surfactant defect. Pediatr Dev Pathol. 2000;3(4):335–345. doi: 10.1007/s100249910048. [DOI] [PubMed] [Google Scholar]
- 49.Corti M, Brody AR, Harrison JH. Isolation and primary culture of murine alveolar type II cells. Am J Respir Cell Mol Biol. 1996;14(4):309–315. doi: 10.1165/ajrcmb.14.4.8600933. [DOI] [PubMed] [Google Scholar]
- 50.Lawrence M, et al. Software for computing and annotating genomic ranges. PLoS Comput Biol. 2013;9(8):e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Olumi AF. Commentary on “integrative genomic analyses reveal an androgen-driven somatic alteration landscape in early-onset prostate cancer.” Weischenfeldt J, Simon R, Feuerbach L, Schlangen K, Weichenhan D, Minner S, Wuttig D, Warnatz HJ, Stehr H, Rausch T, Jäger N, Gu L, Bogatyrova O, Stütz AM, Claus R, Eils J, Eils R, Gerhäuser C, Huang PH, Hutter B, Kabbe R, Lawerenz C, Radomski S, Bartholomae CC, Fälth M, Gade S, Schmidt M, Amschler N, Haß T, Galal R, Gjoni J, Kuner R, Baer C, Masser S, von Kalle C, Zichner T, Benes V, Raeder B, Mader M, Amstislavskiy V, Avci M, Lehrach H, Parkhomchuk D, Sultan M, Burkhardt L, Graefen M, Huland H, Kluth M, Krohn A, Sirma H, Stumm L, Steurer S, Grupp K, Sültmann H, Sauter G, Plass C, Brors B, Yaspo ML, Korbel JO, Schlomm T, Genome Biology Unit, European Molecular Biology Laboratory (EMBL), Heidelberg, Germany.: Cancer Cell 2013;23(2):159-70. Urol Oncol. 2014;32(2):212. doi: 10.1016/j.ccr.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 52.Dautel SE, et al. Lipidomics reveals dramatic lipid compositional changes in the maturing postnatal lung. Sci Rep. 2017;7:40555. doi: 10.1038/srep40555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 54.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37(Web Server issue):W305–W311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37(8):911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 56.Mason RJ, Nellenbogen J, Clements JA. Isolation of disaturated phosphatidylcholine with osmium tetroxide. J Lipid Res. 1976;17(3):281–284. [PubMed] [Google Scholar]
- 57.Yu LM, et al. Constrained sessile drop as a new configuration to measure low surface tension in lung surfactant systems. J Appl Physiol. 2004;97(2):704–715. doi: 10.1152/japplphysiol.00089.2003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.