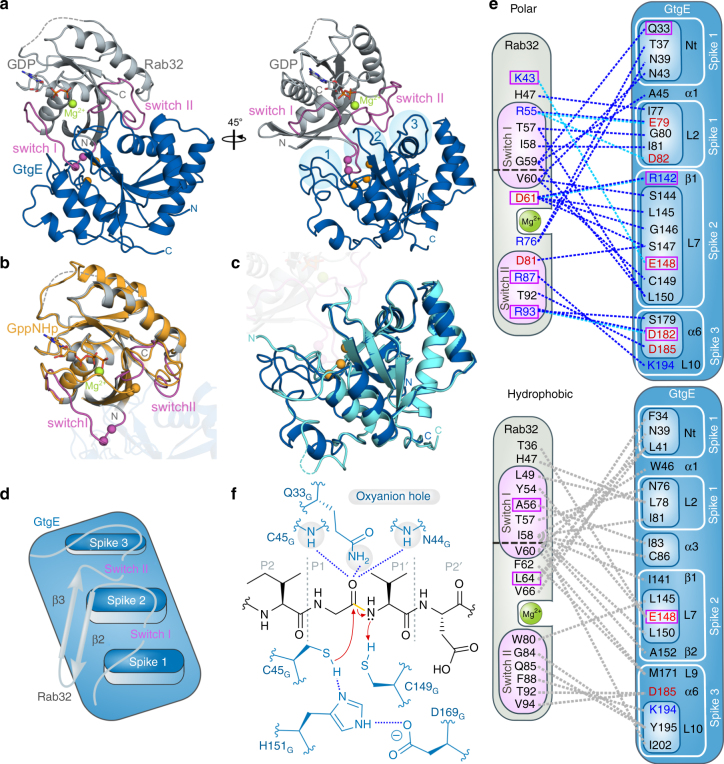
Fig. 3.
Rab32:GDP:GtgEC45A complex structure. a Cartoon depiction of the Rab32:GDP:GtgEC45A-complex. Sticks: GDP; magenta: switch regions; magenta spheres: cleavage site, green sphere: Mg2+-ion; orange spheres: catalytic triad; light blue circles: spike 1–3. b Structural superposition of Rab32:GppCH2p (PDB ID: 4CYM, wheat17) and Rab32:GDP:GtgEC45A (gray). Transparent: GtgE structure. For labeling see panel a. c Structural superposition of GtgE (PDB ID: 5KDG, light blue8) and Rab32:GDP:GtgEC45A.(blue). Transparent: Rab32:GDP structure. For labeling see panel a. d Schematic model of the Rab32:GtgE-binding interface, including the Rab32 switch regions and the adjacent secondary structure elements. The switch regions are positioned into the cavities established by GtgEs spike 1/spike 2 and spike 2/spike 3. e Schematic representation of all polar (top) and hydrophobic interactions (bottom). Acidic and basic amino acids are depicted in red and blue, respectively (Supplementary Table 2). Interactions highlighted in dashed lines: hydrogen bonds (blue), salt-bridges (cyan blue), hydrophobic (gray). L: loop, α: α-helix, β: β-sheet (Supplementary Fig. 1). Magenta box: residues tested by mutations in a GtgE-mediated cleavage assay (Fig. 4c, d). f Schematic of the catalytic mechanism of GtgE. Cleavage occurs in switch I between G59R (P1) and V60R (P1′) by the catalytic triad (C45G, H151G, D169G). Q33G, C45G, N44G: oxyanion hole; proton donor: C149G. Blue: GtgE amino acids, yellow: cleaved bond, bluedashed lines: polar interactions. P1, P2, P1′, and P2′ correspond to the Schechter-Berger-nomenclature of protease substrates