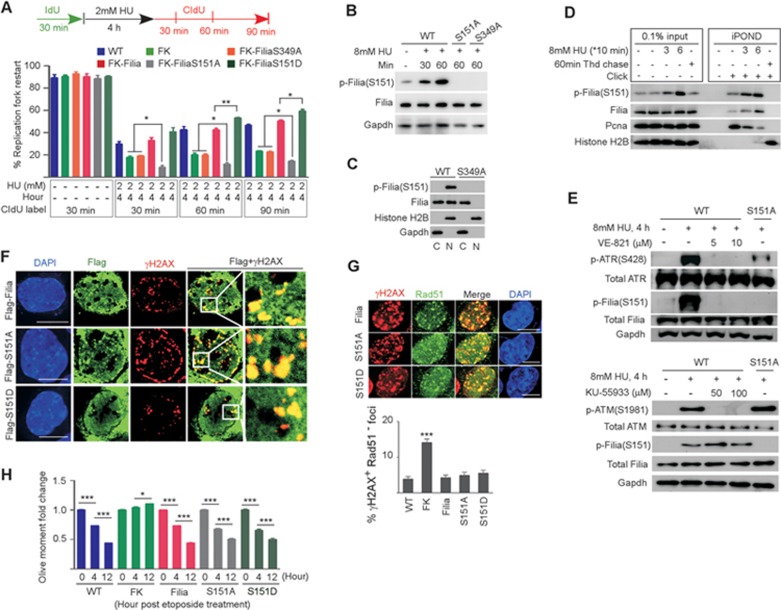
Figure 4.
ATR-dependent phosphorylation of the S151 residue on Filia is required for the restart of replication forks. (A) Rates of replication fork restart in WT, Filia-knockout (FK), FiliaS349A-rescued (FK-FiliaS349A), Filia-rescued (FK-Filia), FiliaS151A-rescued (FK-FiliaS151A) and FiliaS151D-rescued (FK-FiliaS151D) ESCs. (B) Phosphorylation of S151 on Filia (p-(FiliaS151)) was induced by HU treatment in WT ESCs, and no signal was detected in FiliaS151A- or FiliaS349A-complemented ESCs. (C) p-Filia(S151) was present in the nuclear fraction (N) but not the cytoplasmic fraction (C) of the WT ESCs. (D) iPOND confirmed that p-Filia(S151) localized at replication forks under normal and HU-treated conditions in ESCs. (E) Inhibition of the ATR kinase activity in ESCs suppressed the phosphorylation of FiliaS151 in response to HU treatment (upper panel), whereas inhibition of ATM kinase activity had no effect (lower panel). (F) Upon etoposide treatment of ESCs, Flag-tagged Filia, FiliaS151A and FiliaS151D were equally efficient at relocalizing to DNA damage sites. (G) Compared to WT ESCs, Filia-knockout (FK) ESCs were impaired in recruiting Rad51 proteins to DNA damage sites. WT Filia (Filia), FiliaS151A (S151A) and FiliaS151D (S151D) displayed equal ability in rescuing this defect. (H) Filia-knockout (FK) ESCs displayed lower DNA repair ability than WT cells after etoposide treatment. WT Filia, FiliaS151A and FiliaS151D mutants had a similar ability to rescue this defect. Data are represented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001. Scale bar, 10 μm.