FIG 4.
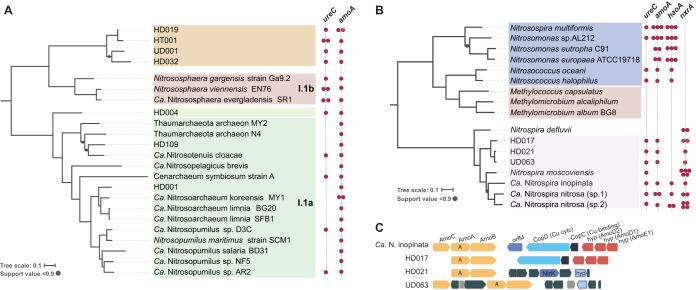
Recovery of indigenous archaeal and bacterial ammonia-oxidizing populations. (A and B) Phylogenetic reconstruction of archaeal (A) and bacterial (B) MAGs harboring amoA genes. Concatenated alignments of conserved genes for bacterial or archaeal genomes were used to build maximum-likelihood trees in RAxML. Colored circles on the right of each tree show the presence of selected nitrification genes. Strikethrough circles indicate incomplete sequences detected in MAGs. (C) Comparison of the genomic context for the amoA genes found in “Ca. Nitrospira inopinata” and nitrifier bacterial MAGs recovered from both sites. Colors denote different gene operons.